Patients
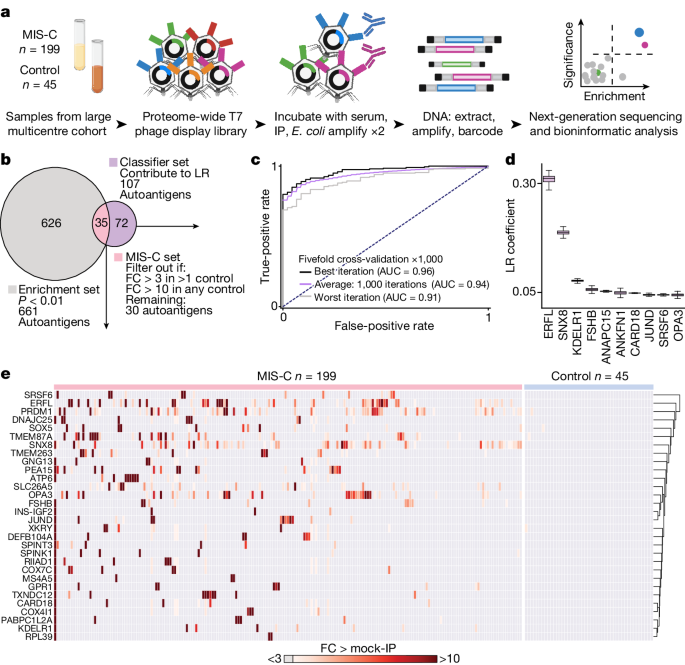
Patients were recruited through the prospectively enrolling multicentre Overcoming COVID-19 and Taking on COVID-19 Together study in the USA. All patients meeting clinical criteria were included in the study, and therefore no statistical methods were used to predetermine sample size and no blinding or randomization of subjects occurred. The study was approved by the central Boston Childrenâs Hospital Institutional Review Board (IRB) and reviewed by IRBs of participating sites with CDC IRB reliance. A total of 292 patients consented and were enrolled into one of the following independent cohorts between 1 June 2020 and 9 September 2021: 223 patients hospitalized with MIS-C (199 in the primary discovery cohort and 24 in a separate subsequent validation cohort), 29 patients hospitalized for COVID-19 in either an intensive care or step-down unit (referred to as âsevere acute COVID-19â in this study) and 45 outpatients (referred to as âat-risk controlsâ in this study) post-SARS-CoV-2 infections associated with mild or no symptoms. The demographic and clinical data are summarized in Extended Data Tables 1â3. The 2020 US CDC case definition was used to define MIS-C51. All patients with MIS-C had positive SARS-CoV-2 serology results and/or positive SARS-CoV-2 test results by reverse transcriptase quantitative PCR. All patients with severe COVID-19 or outpatient SARS-CoV-2 infections had a positive antigen test or nucleic acid amplification test for SARS-CoV-2. For outpatients, samples were collected from 36 to 190 days after the positive test (median of 70 days after a positive test; interquartile range of 56â81 days). For use as controls in the SARS-CoV-2-specific PhIP-seq, plasma from 48 healthy, pre-COVID-19 controls were obtained as deidentified samples from the New York Blood Center. These samples were part of retention tubes collected at the time of blood donations from volunteer donors who provided informed consent for their samples to be used for research.
DNA oligomers for SLBAs
DNA coding for the desired peptides for use in SLBAs were inserted into split luciferase constructs containing a terminal HiBiT tag and synthesized (Twist Biosciences) as DNA oligomers and verified by Twist Biosciences before shipment. Constructs were amplified by PCR using the 5â²- AAGCAGAGCTCGTTTAGTGAACCGTCAGA-3â² and 5â²-GGCCGGCCGTTTAAACGCTGATCTT-3â² primer pair.
For SNX8, the oligomers coded for the following sequences:
EADPPASDLPTPQAIEPQAIVQQVPAPSRMQMPQGNPLLLSHTLQELLA
AAAAAAAAAATPQAIEPQAIVQQVPAPSRMQMPQGNPLLLSHTLQELLA
EADPPAAAAAAAAAAEPQAIVQQVPAPSRMQMPQGNPLLLSHTLQELLA
EADPPASDLPAAAAAAAAAAVQQVPAPSRMQMPQGNPLLLSHTLQELLA
EADPPASDLPTPQAIAAAAAAAAAAAPSRMQMPQGNPLLLSHTLQELLA
EADPPASDLPTPQAIEPQAIAAAAAAAAAAQMPQGNPLLLSHTLQELLA
EADPPASDLPTPQAIEPQAIVQQVPAAAAAAAAAANPLLLSHTLQELLA
EADPPASDLPTPQAIEPQAIVQQVPAPSRMAAAAAAAAAASHTLQELLA
EADPPASDLPTPQAIEPQAIVQQVPAPSRMQMPQGAAAAAAAAAAELLA
EADPPASDLPTPQAIEPQAIVQQVPAPSRMQMPQGNPLLLAAAAAAAAA
For SARS-CoV-2 nucleocapsid protein, the oligomers coded for the following sequences:
ATEGALNTPKDHIGTRNPANNAAIVLQLPQGTTLPKGFYAEGSRGGSQA
AAAAAAAAAADHIGTRNPANNAAIVLQLPQGTTLPKGFYAEGSRGGSQA
ATEGAAAAAAAAAAARNPANNAAIVLQLPQGTTLPKGFYAEGSRGGSQA
ATEGALNTPKAAAAAAAAAANAAIVLQLPQGTTLPKGFYAEGSRGGSQA
ATEGALNTPKDHIGTAAAAAAAAAALQLPQGTTLPKGFYAEGSRGGSQA
ATEGALNTPKDHIGTRNPANAAAAAAAAAAGTTLPKGFYAEGSRGGSQA
ATEGALNTPKDHIGTRNPANNAAIVAAAAAAAAAAKGFYAEGSRGGSQA
ATEGALNTPKDHIGTRNPANNAAIVLQLPQAAAAAAAAAAEGSRGGSQA
ATEGALNTPKDHIGTRNPANNAAIVLQLPQGTTLPAAAAAAAAAAGSQA
ATEGALNTPKDHIGTRNPANNAAIVLQLPQGTTLPKGFYAAAAAAAAAA
DNA plasmids for RLBAs
For RLBAs, DNA expression plasmids under control of a T7 promoter and with a terminal MycâDDK tag for the desired protein were utilized. For ERFL, a custom plasmid was ordered from Twist Bioscience in which a MycâDDK-tagged full-length ERFL sequence under a T7 promoter was inserted into the pTwist Kan High Copy Vector (Twist Bioscience). Twist Bioscience verified a sequence-perfect clone by next-generation sequencing before shipment. Upon receipt, the plasmid was sequence verified by Primordium Labs. For SNX8, a plasmid containing the MycâDDK-tagged full-length human SNX8 under a T7 promoter was ordered from Origene (RC205847) and was sequence verified by Primordium Labs upon receipt. For KDELR1, a plasmid containing the MycâDDK-tagged full-length human KDELR1 under a T7 promoter was ordered from Origene (RC205880) and was sequence verified by Primordium Labs upon receipt. For IL1RN, a plasmid containing the MycâDDK-tagged full-length human IL1RN under a T7 promoter was ordered from Origene (RC218518) and was sequence verified by Primordium Labs upon receipt.
Polypeptide pools for activation-induced marker assays
To obtain polypeptides tiling the full-length SNX8 protein, 15-mer polypeptide fragments with 11-amino acid overlaps were ordered from JPT Peptide Technologies and synthesized. Together, a pool of 130 of these polypeptides (referred to as the âSNX8 poolâ) spanned all known translated SNX8 (the full-length 465-amino acid SNX8 protein, as well as a unique region of SNX8 isoform 3). A separate pool was designed to cover primarily the region of SNX8 with similarity to the SARS-CoV-2 nucleocapsid protein in high resolution (referred to as the âhigh-resolution epitope poolâ). This pool contained 20 10-mers with 9-amino acid overlaps tiling amino acids 44â72 (IVQQVPAPSRMQMPQGNPLLLSHTLQELL) of the full-length SNX8 protein. The sequence of each of these 150 polypeptides was verified by mass spectrometry and purity was calculated by high-performance liquid chromatography (HPLC).
Peptides for tetramer assays
For use in loading tetramers, three peptides were ordered from Genemed Synthesis as 9-mers. LQLPQGTTL and LQLPQGITL correspond to the region of the SARS-CoV-2 nucleocapsid protein with similarity to human SNX8 in the ancestral sequence and a minor variant, respectively. This sequence was verified by mass spectrometry and purity was calculated as 96.61% by HPLC. The other sequence, MQMPQGNPL, corresponds to the region of human SNX8 protein with similarity to the SARS-CoV-2 nucleocapsid protein. This sequence was verified by mass spectrometry and purity was calculated as 95.83% by HPLC.
Human proteome PhIP-seq
Human proteome PhIP-seq was performed following our previously published vacuum-based PhIP-seq protocol12 (https://www.protocols.io/view/scaled-high-throughput-vacuum-phip-protocol-ewov1459kvr2/v1).
Our human peptidome library consists of a custom-designed phage library of 731,724 unique T7 bacteriophage each presenting a different 49-amino acid peptide on its surface. Collectively, these peptides tile the entire human proteome including all known isoforms (as of 2016) with 25-amino acid overlaps. Of the phage library, 1âml was incubated with 1âμl of human serum overnight at 4â°C and immunoprecipitated with 25âμl of 1:1 mixed protein A and protein G magnetic beads (10008D and 10009D, Thermo Fisher). These beads were than washed, and the remaining phageâantibody complexes were eluted in 1âml of Escherichia coli (BLT5403, EMD Millipore) at 0.5â0.7âOD and amplified by growing in a 37â°C incubator. This new phage library was then re-incubated with the serum from the same individual and the previously described protocol was repeated. DNA was then extracted from the final phage library, barcoded, PCR amplified and Illumina adaptors were added. Next-generation sequencing was performed using an Illumina sequencer (Illumina) to a read depth of approximately 1 million per sample.
Human proteome PhIP-seq analysis
All human peptidome analysis (except when specifically stated otherwise) was performed at the gene level, in which all reads for all peptides mapping to the same gene were summed, and 0.5 reads were added to each gene to allow inclusion of genes with zero reads in mathematical analyses. Within each individual sample, reads were normalized by converting to the percentage of total reads. To normalize each sample against background nonspecific binding, a fold change over mock-IP was calculated by dividing the sample read percentage for each gene by the mean read percentage of the same gene for the AG bead-only controls. This fold-change signal was then used for side-by-side comparison between samples and cohorts. Fold-change values were also used to calculate z scores for each patient with MIS-C compared with controls and for each control sample by using all remaining controls. These z scores were used for the logistic-regression feature weighting. In instances of peptide-level analysis, raw reads were normalized by calculating the number of reads per 100,000 reads.
SARS-CoV-2 proteome PhIP-seq
SARS-CoV-2 proteome PhIP-seq was performed as previously described39. In brief, 38 amino acid fragments tiling all open reading frames from SARS-CoV-2, SARS-CoV-1 and 7 other CoVs were expressed on T7 bacteriophage with 19-amino acid overlaps. Of the phage library, 1âml was incubated with 1âμl of human serum overnight at 4â°C and immunoprecipitated with 25âμl of 1:1 mixed protein A and protein G magnetic beads (10008D and 10009D, Thermo Fisher). Beads were washed five times on a magnetic plate using a P1000 multichannel pipette. The remaining phageâantibody complexes were eluted in 1âml of E. coli (BLT5403, EMD Millipore) at 0.5â0.7âOD and amplified by growing in 37â°C incubator. This new phage library was then re-incubated with the serum of the same individual and the previously described protocol was repeated for a total of three rounds of immunoprecipitations. DNA was then extracted from the final phage library, barcoded, PCR amplified and Illumina adaptors were added. Next-generation sequencing was then performed using an Illumina sequencer (Illumina) to a read depth of approximately 1 million per sample.
Coronavirus proteome PhIP-seq analysis
To account for differing read depths between samples, the total number of reads for each peptide fragment was converted to the number of reads per 100,000 (RPK). To calculate normalized enrichment relative to pre-COVID-19 controls (FCâ>âpre-COVID-19), the RPK for each peptide fragment within each sample was divided by the mean RPK of each peptide fragment among all pre-COVID-19 controls. These FCâ>âpre-COVID-19 values were used for all subsequent analyses as described in the text and figures.
RLBA
RLBAs were performed as previously described12,32. In brief, DNA plasmids containing full-length cDNA under the control of a T7 promoter for each of the validated antigens (see âDNA plasmids for RLBAsâ above) were verified by Primordium Labs sequencing. The respective DNA templates were used in the T7 TNT in vitro transcription/translation kit (L1170, Promega) using [35S]-methionine (NEG709A, PerkinElmer). Respective protein was column purified on Nap-5 columns (17-0853-01, GE Healthcare), and equal amounts of protein (approximately 35,000 counts per minute) were incubated overnight at 4â°C with 2.5âμl of serum or 1âμl of anti-Myc-positive control antibody (1:10 dilution; 2272S, Cell Signaling Technology). Immunoprecipitation was then performed on 25âμl of Sephadex protein A/G beads (4:1 ratio; GE17-5280-02 and GE17-0618-05, Sigma-Aldrich) in 96-well polyvinylidene difluoride filtration plates (EK-680860, Corning). After thoroughly washing, the counts per minute of immunoprecipitated protein was quantified using a 96-well Microbeta Trilux liquid scintillation plate reader (Perkin Elmer).
SLBA
SLBA was performed as previously described52. A detailed SLBA protocol is available on protocols.io (https://doi.org/10.17504/protocols.io.4r3l27b9pg1y/v1).
In brief, the DNA oligomers listed above (see âDNA oligomers for SLBAsâ) were amplified by PCR using the primer pairs listed above (see âDNA oligomers for SLBAsâ). Unpurified PCR product was used as input in the T7 TNT in vitro transcription/translation kit (L1170, Promega) and the Nano-Glo HiBit Lytic Detection System (N3040, Promega) was used to measure relative luciferase units of translated peptides in a luminometre. Equal amounts of protein (in the range of 2âÃâ106â2âÃâ107 relative luciferase units) were incubated overnight with 2.5âμl patient sera or 1âμl anti-HiBit-positive control antibody (1:10 dilution; CS2006A01, Promega) at 4â°C. Immunoprecipitation was then performed on 25âµl of Sephadex protein A/G beads (1:1 ratio; GE17-5280-02 and GE17-0618-05, Sigma-Aldrich) in 96-well polyvinylidene difluoride filtration plates (EK-680860, Corning). After thoroughly washing, luminescence was measured using the Nano-Glo HiBit Lytic Detection System (N3040, Promega) in a luminometre.
Activation-induced marker assay
PBMCs were obtained from ten patients with MIS-C and ten controls for use in the activation-induced marker assay. PBMCs were thawed, washed, resuspended in serum-free RPMI medium and plated at a concentration of 1âÃâ106 cells per well in a 96-well round-bottom plate. For each individual, PBMCs were stimulated for 24âh with either the SNX8 pool (see above) at a final concentration of 1âmgâmlâ1 per peptide in 0.2% DMSO or a vehicle control containing 0.2% DMSO only. For four of the controls and two of the patients with MIS-C, there were sufficient PBMCs for an additional stimulation condition using the SNX8 high-resolution epitope pool (see above) also at a concentration of 1âmgâmlâ1 per peptide in 0.2% DMSO for 24âh. Following the stimulation, cells were washed with FACS buffer (Dulbeccoâs PBS without calcium or magnesium, 0.1% sodium azide, 2âmM EDTA and 1% FBS) and stained with the following antibody panel each at 1:100 dilution for 20âmin at 4â°C, and then flow cytometry analysis was immediately performed.
For the antibody panel: anti-CD3 Alexa 647 (clone OKT3, 317312, BioLegend), anti-CD4 Alexa 488 (clone OKT4, 317420, BioLegend), anti-CD8 Alexa 700 (clone SK1, 344724, BioLegend), anti-OX-40 (also known as CD134) PE-Dazzle 594 (clone ACT35, 350020, BioLegend), anti-CD69 PE (clone FN-50, 310906, BioLegend), anti-CD137 (also known as 4-1BB) BV421 (clone 4B4-1, 309820, BioLegend), anti-CD14 PerCP-Cy5 (clone HCD14, 325622, BioLegend), anti-CD16 PerCP-Cy5 (clone B73.1, 360712, BioLegend), anti-CD19 PerCP-Cy5 (clone HIB19, 302230, BioLegend) and Live/Dead Dye eFluor 506 (65-0866-14, Invitrogen).
The activation-induced marker analysis was performed using FlowJo software using the gating strategy shown in Extended Data Fig. 7a. All gates were fixed within each condition of each sample. Activated CD4 T cells were defined as those that were co-positive for OX40 and CD137. Activated CD8 T cells were defined as those that were co-positive for CD69 and CD137. Gating thresholds for activation were defined by the outer limits of signal in the vehicle controls allowing for up to two outlier cells. Frequencies were calculated as a percentage of total CD3+ cells (T cells). Two MIS-C samples had insufficient total events captured by flow cytometry (total of 5,099 and 4,919 events, respectively) and were therefore removed from analysis.
Initial tetramer assay
For the initial tetramer assay, see Extended Data Fig. 4a. PBMCs from two patients with MIS-C with HLA-A*02:01 (HLA typed from PAXgene RNAseq, one confirmed by serotyping), one patient with MIS-C with HLA-B*35:01 (HLA typed from PAXgene RNAseq) and three at-risk controls with HLA-A*02.01 (all three identified by serotyping, two of three confirmed by PAXgene RNAseq HLA typing; the other sample did not have genomic DNA available for genotyping) were thawed, washed and put into culture with media containing recombinant human IL-2 at 10ângâmlâ1 in 96-well plates. The peptide fragments (details above) LQLPQGITL and MQMPQGNPL were then added to PBMCs to a final concentration of 10âmgâmlâ1 per peptide and incubated (37â°C at 5% CO2) for 7 days.
Following the 7 days of incubation, a total of eight pHLA class I tetramers were generated from UV-photolabile biotinylated monomers, four each from HLA-A*02:01 and HLA-B*35:01 (NIH Tetramer Core). Peptides were loaded via UV peptide exchange. Tetramerization was carried out using streptavidin conjugated to fluorophores PE and APC or BV421 followed by quenching with 500âµM d-biotin, similar to our previously published methods44,53. Tetramers were then pooled together as shown below:
For the HLA-A*02:01 pool, the MADS (LQLPQGITL)-loaded PE tetramer, MADS (LQLPQGITL)-loaded APC tetramer, SNX8 (MQMPQGNPL)-loaded PE tetramer and SNX8 (MQMPQGNPL)-loaded BV421 tetramer were used, all with HLA-A*02:01 restriction.
For the HLA-B*35:01 pool, the MADS (LQLPQGITL)-loaded PE tetramer, MADS (LQLPQGITL)-loaded APC tetramer, SNX8 (MQMPQGNPL)-loaded PE tetramer and SNX8 (MQMPQGNPL)-loaded BV421 tetramer were used, all with HLA-B*35:01 restriction.
All PBMCs were then treated with 100ânM dasatinib (StemCell) for 30âmin at 37â°C followed by staining (no wash step) with the respective tetramer pool corresponding to their HLA restriction (final concentration of 2â3âµgâmlâ1) for 30âmin at 25â°C. Cells were then stained with the following cell-surface markers each at 1:100 dilution for 20âmin, followed by immediate analysis on a flow cytometer.
For the surface markers: anti-CD8 Alexa 700 (clone SK1, 357404, BioLegend), anti-CD4 PerCP-Cy5 (clone SK1, 300530, BioLegend), anti-CD14 PerCP-Cy5 (clone HCD14, 325622, BioLegend), anti-CD16 PerCP-Cy5 (clone B73.1, 360712, BioLegend), anti-CD19 PerCP-Cy5 (clone HIB19, 302230, BioLegend) and Live/Dead Dye eFluor 506 (65-0866-14, Invitrogen). Streptavidin was conjugated to PE (S866, Invitrogen), APC (S868, Invitrogen) and BV421 (405225, BioLegend).
The gating strategy is outlined in Extended Data Fig. 7b. A stringent tetramer gating strategy was used to identify cross-reactive T cells, in which CD8+ T cells were required to be triple positive for PE, APC and BV421 labels (that is, a single CD8 T cell bound to PE-conjugated LQLPQGITL and/or PE-conjugated MQMPQGNPL in addition to APC-conjugated LQLPQGITL and BV421-conjugated MQMPQGNPL).
Serotyping was performed using an anti-HLA-A2 antibody (1:100 dilution; FITC anti-human HLA-A2 antibody, clone BB7.2, 343303, BioLegend), and pertinent results are shown in Extended Data Fig. 7c.
Assembly of easYmer monomers and fold testing
For the assembly of HLA class I pHLA easYmer monomers and fold testing, see Fig. 4. Unfolded, biotinylated easYmer monomers (Immudex) were obtained for HLA-A*02:01 and HLA-A*02:06. SARS-CoV-2 MADS (LQLPQGITL), SARS-CoV-2 Wuhan (LQLPQGTTL) and human SNX8 (MQMPQGNPL) peptides were commercially synthesized (Genscript), diluted to 1âmM in ddH2O or DMSO, and loaded onto each easYmer allele according to the manufacturerâs instructions at 18â°C for 48âh. Proper pHLA monomer formation and MADS and SNX8 peptide-binding strength were evaluated for each HLA using a âβ2m fold testâ relative to negative (no peptide; unloaded monomer) and positive (strong binding peptide; CMV pp65 495â503 (NLVPMVATV)) controls as per the manufacturerâs protocol. In brief, peptide-loaded monomers with a concentration of 500ânM were serially diluted to 9ânM, 3ânM and 1ânM in dilution buffer (1à PBS with 5% glycerol; G5516, Sigma-Aldrich) and incubated with streptavidin beads (6â8âμm; SVP-60-5, Spherotech) at 37â°C for 1âh to allow binding of stable complexes to beads, then washed three times with FACS buffer (1à PBS, 0.5% BSA (A7030, Sigma-Aldrich) and 2âmM EDTA (15575-038, Thermo Fisher Scientific)). Samples were then stained with PE-conjugated anti-human β2m antibody (clone BBM.1, sc-13565, Santa Cruz Biotech) at 1:200 for 30âmin at 4â°C, washed three times with FACS buffer and analysed on a 5 Laser 16UV-16V-14B-10YG-8R AURORA spectral cytometer (Cytek). pHLA-binding strength positively correlated with stability and concentration of the pHLAâβ2m complex. Therefore, the geometric mean fluorescence intensity of anti-β2m staining in this assay reports on the strength of the pHLA binding compared with the positive and negative controls. We classified binding strength for each HLA and peptide combination based on the fold change in anti-β2m geometric mean fluorescence intensity over the no-peptide negative control at 9ânM. Strong binders were defined at more than 10-fold higher, moderate binders at more than 3-fold, weak binders at more than 1.5-fold and non-binders at less than 1.5-fold change over the negative control. Flow cytometry data were analysed using FlowJo version 10.7.2 software (BD Biosciences).
pHLA tetramer assembly
For the pHLA tetramer assembly, see Fig. 4. pHLA tetramers were assembled from HLA-A*02:01 and HLA-A*02:06 easYmer monomers (Immudex) with confirmed peptide binding to SARS-CoV-2 MADS (LQLPQGITL), Wuhan (LQLPQGTTL) and SNX8 (MQMPQGNPL) peptides according to the manufacturerâs instructions. In brief, fluorochrome-conjugated streptavidin (0.2âmgâmlâ1, PE, 405203, BioLegend; 0.2âmgâmlâ1, APC, 405207, BioLegend; and BV421, 405226, BioLegend) was added to loaded monomers at 8âng per 1âμl pHLA complex (500ânM) in three volumes. After each 1/3 volume addition, samples were mixed and incubated for 15âmin at 4â°C in the dark. Assembled tetramers were stored at 4â°C in the dark until use.
Enhanced peptide-specific T cell expansion
For enhanced peptide-specific T cell expansion, see Fig. 4. PBMCs from MIS-C confirmed participants with HLA-A*02:01 or HLA-A*02:06 were obtained for peptide-specific expansion according to published methods54 before single-cell sorting of tetramer-positive T cells. On expansion day 0, PBMCs were thawed, counted and seeded onto 96-well round-bottom plates at 100,000 cells per well in 200âμl antigen-presenting cell differentiation media (X-VIVO 15 serum-free haematopoietic cell medium (04-418Q, Lonza) supplemented with human GM-CSF (1,000âIUâmlâ1; 130-095-372, Miltenyi Biotec), human IL-4 (500âIUâmlâ1; 204-IL-010, R&D Systems) and human Flt3-L (50ângâmlâ1; 308-FKN-025, R&D Systems) final concentrations) and incubated for 24âh at 37â°C and 5% CO2. On day 1, 100âμl cell supernatant was replaced with 100âμl Adjuvant Solution (X-VIVO 15 supplemented with R848 (10âμM; tlrl-r848-5, InvivoGen), lipopolysaccharide (Salmonella minnesota; 100ângâmlâ1; tlrl-smlps, InvivoGen) and human IL-1β (10ângâmlâ1; 201-LB-010, R&D Systems) final concentrations) and pooled MADS (LQLPQGITL) and SNX8 (MQMPQGNPL) peptides at a final concentration of 10âμM each. No-peptide control wells were set up for each sample by adding a 1:2 dilution of DMSO in H2O to match the peptide volume and diluent. Cells were incubated for 24âh at 37â°C and 5% CO2. On days 2, 4, 7 and 9, 100âμl supernatant was replaced with 100âμl T cell expansion solution: RP-10 (RPMI 1640 (22400-089, Gibco), 10% heat-inactivated human serum AB (100-512, Gemini Bio-Products), 10âmM HEPES, 0.1âmgâmlâ1 gentamicin (15750-060, Thermo Fisher Scientific) and 1à GlutaMAX (35050-061, Gibco)) supplemented with human IL-2 (10âIUâmlâ1; 202-IL-050, R&D Systems), human IL-7 (10ângâmlâ1; 207-IL-025, R&D Systems) and human IL-15 (10ângâmlâ1; 200-15, PeproTech) final concentrations. On day 10, peptide-expanded cells from an individual participant were pooled; cells from no-peptide controls were collected separately.
Single-cell index sorting
Unexpanded PBMCs (direct ex vivo) or peptide-expanded T cells were obtained, washed in 1à PBS and treated with 100ânM dasatinib (CDS023389, Sigma-Aldrich) in 1à PBS for 30âmin at 37â°C and 5% CO2 (ref. 55). Cells were then pelleted and resuspended in 50âμl FACS buffer (1à PBS and 0.04% BSA) supplemented with human TruStain FcX blocking buffer (1:10 dilution; 422302, BioLegend), 500âμM d-biotin (B20656, Thermo Fisher Scientific) and a unique tetramer cocktail containing MADSâtetramerâPE (1:10 dilution), MADSâtetramerâAPC (1:10 dilution), SNX8âtetramerâPE (1:10 dilution) and SNX8âtetramerâBV421 (1:10 dilution) based on participant HLA type (A*02:01 and A*02:06). Cells were incubated in the dark at 25â°C for 1âh followed by direct addition of 50âμl (100âμl total volume) of FACS supplemented with 500âμM d-biotin and an antibody cocktail containing FITC-conjugated anti-human CD3 (1:20 dilution; clone OKT3, lot B390808, 317306, BioLegend), BV605-conjugated anti-human CD8 (1:20 dilution; clone SK1, lot B371925, 344742, BioLegend), BV510-conjugated anti-human CD4 (1:20 dilution; clone OKT4, lot B375526, 317444, BioLegend), BV510-conjugated anti-human CD14 (1:20 dilution; clone 63D3, lot B390770, 367124, BioLegend), BV510-conjugated anti-human CD16 (1:20 dilution; clone 3G8, lot B372132, 302048, BioLegend), BV510-conjugated anti-human CD19 (1:20 dilution; clone HIB19, lot B390665, 302242, BioLegend) and Ghost Dye Violet 510 Viability Dye (1:400 dilution; lot D0870061322133, 13-0870-T500, Tonbo Biosciences) for 30âmin in the dark at 4â°C. Cells were then pelleted, washed twice with 4âml FACS buffer (containing 500âμM d-biotin), suspended in 500âμl FACS (containing 500âμM d-biotin) and passed through a 45-μM filter before proceeding to single-cell sorting on a Sony SY3200 cell sorter. Individual, live, BV510 dump gate (CD4, CD14, CD16 and CD19)-negative, CD3+CD8+ T lymphocytes were gated to distinguish tetramer triple-positive cells (PE+APC+BV421+) as described in Extended Data Fig. 7d and sorted into individual wells of a 384-well plate loaded with Superscript VILO master mix (11754250, Thermo Fisher Scientific). After sorting, plates were centrifuged at 500g and stored at â80â°C until processing.
Paired TCRαβ amplification and sequencing
Single-cell paired TCRα and TCRβ chain library preparation and sequencing was performed on T cells sorted into 384-well index plates as previously described56. In brief, after reverse transcription of cells sorted in Superscript VILO master mix, cDNA underwent two rounds of nested multiplex PCR amplification using a mix of human V-segment-specific forward primers and human TRAC and TRBC segment-specific reverse primers (see Supplementary Table 1 for primer details). Resulting TCRα and TCRβ amplicons were sequenced on an Illumina MiSeq at 2âÃâ150-bp read length.
Cell lines
All cultured cell lines were maintained at 37â°C and 5% CO2 in a humidified incubator. HEK 293T cells (CRL-3216, American Type Culture Collection) were purchased from the American Type Culture Collection and verified commercially. HEK 293T cells were cultured in DMEM (11965-092, Gibco) supplemented with 10% FBS (16140-071, Gibco), 2âmM l-glutamine (25030-081, Gibco) and 100âUâmlâ1 penicillinâstreptomycin (15140-122, Gibco). 2D3 Jurkat J76.7 cells57,58 (TCR-null, CD8+) expressing an NFATâeGFP reporter were kindly provided by F. Fujiki and were cultured in RPMI 1640 (22400-089, Gibco) supplemented with 10% FBS, 2âmM l-glutamine and 100âUâmlâ1 penicillinâstreptomycin. All cell lines were confirmed to be mycoplasma negative during the course of experiments.
TCR repertoire analysis
TCR similarity networks were constructed as previously described49,59. In brief, to measure the distance between TCRαβ clonotypes, we used the TCRdist algorithm implementation from the CoNGA v0.1.2 Python package47. Further analysis was performed using the R language for statistical computing, with merging and subsetting of data performed using the dplyr v1.1.4 package. TCR similarity networks were built using stringdist v0.9.12 and igraph v2.0.3 (ref. 60) R packages, and visualized using gephi v0.9.7 (ref. 61) software.
TCR reconstruction and cloning
Full-length TCRαβ sequences were reconstructed from V/J gene usage and CDR3 sequences using Stitchr v1.0.0 (ref. 62) for each index-sorted T cell. TCRα and TCRβ chain sequences were modified to use murine constant regions and joined by a 2A element from thosea asigna virus (T2A). A sequence encoding mCherry was additionally appended by a 2A element from porcine teschovirus (P2A) as a fluorescent marker of transduction. The full-length gene fragment encoding TCRβâT2AâTCRαâP2AâmCherry was synthesized and cloned commercially (Genscript) into the lentiviral vector pLVX-EF1α-IRES-Puro (631253, Takara).
Generation of TCR-expressing Jurkat cells
To generate transducing particles packaging individual TCRs of interest (Fig. 4c), HEK 293T cells were transduced with a pLVX lentiviral vector encoding a unique TCRαβâmCherry insert, psPAX2 packaging plasmid (plasmid #12260, Addgene) and an pMD2.G envelope plasmid (plasmid #12259, Addgene) at a ratio of 4:3:1. At 24âh and 48âh post-transfection, viral supernatants were harvested, passed through a 0.45-µm SFCA filter (723-9945, Thermo Fisher Scientific), concentrated using Lenti-X Concentrator (631232, Takara) and stored at â80â°C as single-use aliquots. To generate TCR-expressing Jurkat cell lines (Jurkat-TCR+), 2D3 Jurkat J76.7 cells (TCR-null, CD8+, NFATâeGFP reporter) were seeded in a 12-well tissue-culture-treated plate at 1âÃâ106 cells per well in complete RPMI (RPMI 1640, 10% FBS, 2âmM l-glutamine, 100âUâmlâ1 penicillinâstreptomycin) and transduced by adding concentrated lentivirus dropwise to each well. At 48â72âh post-tranduction, puromycin was added at 1âμgâmlâ1 and cultured for 1 week to select for transduced cells. Jurkat-TCR+ cell lines were validated for the presence of correctly folded TCR on the cell surface by flow cytometry using a monoclonal antibody targeting the mouse TCRβ constant region (APC/Fire750-conjugated; clone H57-597, 109246, BioLegend; Extended Data Fig. 5a). Flow cytometry data were collected on a custom-configured BD Fortessa using FACSDiva software (v8.0.1; Becton Dickinson) and analysed using FlowJo version 10.7.2 software (BD Biosciences).
Specificity validation of putative cross-reactive TCR sequences
The specificity of TCR-expressing Jurkat T cell lines was validated by tetramer staining using the same reagents used for single-cell sorting PBMCs (above). In brief, 1âÃâ106 Jurkat-TCR+ cell lines or untransduced Jurkat J76.7 (TCR-null; background control) were washed in 1à PBS and resuspended in 50âμl FACS buffer (1à PBS and 0.04% BSA) and a unique tetramer cocktail containing MADSâtetramerâPE (1:10 dilution), MADSâtetramerâAPC (1:10 dilution), SNX8âtetramerâPE (1:10 dilution) and SNX8âtetramerâBV421 (1:10 dilution) based on the restricting HLA type (A*02:01 and A*02:06). Tetramers conjugated to the Wuhan peptide sequence (LQLPQGTTL), including WuhanâtetramerâPE (1:10 dilution) and WuhanâtetramerâAPC (1:10 dilution), were also tested. A second set of wells were set up in which each individual tetramer was used to stain cells. Cells were incubated in the dark at 25â°C for 30âmin after which 50âµl of FACS buffer containing Ghost Dye Violet 510 Viability Dye (1:400 dilution; lot D0870061322133, 13-0870-T500, Tonbo Biosciences) was added for an additional 30-min incubation in the dark at 25â°C. Cells were then washed twice with 1âml FACS buffer, suspended in 300âμl FACS and analysed by flow cytometry on a custom-configured BD Fortessa using FACSDiva software (v8.0.1; Becton Dickinson). Cell population gating and fluorescence analysis was performed using FlowJo version 10.7.2 software (BD Biosciences) as described in Extended Data Fig. 7e.
scRNA-seq analysis
To assess the cell-type specificity in a relevant disease context, we analysed SNX8 expression from a single-cell sequencing of PBMC samples from patients with severe, mild or asymptomatic COVID-19 infection, influenza virus infection and healthy controls48. Gene expression data from 59,572 pre-filtered cells were downloaded from the Gene Expression Omnibus database under accession GSE149689 for analysis and downstream processing with scanpy v1.10.0 (ref. 63). Cells with (1) less than 1,000 total counts, (2) less than 800 expressed genes, and (3) more than 3,000 expressed genes were filtered out as further quality control, leaving 42,904 cells for downstream analysis. Gene expression data were normalized to have 10,000 counts per cell and were log1p transformed. Highly variable genes were calculated using the scanpy function highly_variable_genes using Seurat flavor with the default parameters (min_mean = 0.0125, max_mean = 3, and min_disp = 0.5)64. Only highly variable genes were used for further analysis. The total number of counts per cell was regressed out, and the gene expression matrix was scaled using the scanpy function scale with max_valueâ=â10. Dimensionality reduction was performed using principal components analysis with 50 principal components. Batch balanced k-nearest neighbours, implemented with scanpyâs function bbknn, was used to compute the top neighbours and normalize batch effects65. The batch-corrected cells were clustered using the Leiden algorithm and projected into two dimensions with uniform manifold approximation and projection for visualization. Initial cluster identity was determined by finding marker genes with differential expression analysis performed using a Studentâs t-test on log1p-transformed raw counts with the scanpy function rank_genes_groups66,67.
Statistical methods
All statistical analysis was performed in Python using the Scipy Stats package unless otherwise indicated. For comparisons of distributions of PhIP-seq enrichment between two groups, a non-parametric KolmogorovâSmirnov test was utilized. For logistic-regression feature weighting, the Scikit-learn package68 was used, and logistic-regression classifiers were applied to z-scored PhIP-seq values from individuals with MIS-C versus at-risk controls. A liblinear solver was used with L1 regularization, and the model was evaluated using a five-fold cross-validation (four of the five for training, and one of the five for testing). For the RLBAs and SLBAs, first an antibody index was calculated as follows: (sample valueâââmean blank value)/(positive control antibody valuesâââmean blank values). For the alanine mutagenesis scans, blank values of each construct were combined, and a single mean was calculated. A normalization function was then applied to the experimental samples only (excluding antibody-only controls) to create a normalized antibody index ranging from 0 to 1. Comparisons between two groups of samples were performed using a MannâWhitney U-test. An antibody was considered to be âpositiveâ when the normalized antibody index in a sample was greater than 3 s.d. above the mean of controls. When comparing two groups of normally distributed data, a Studentâs t-test was performed.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.