Fly husbandry and stocks
Flies were reared on a standard cornmeal medium at 25 °C under a 12-h–12-h light–dark cycle. To enhance transgene expression levels, flies from all genetic perturbation experiments, including control groups, were shifted to 29 °C shortly before puparium formation. Detailed genotypes for each experiment are listed in Supplementary Table 1.
Molecular cloning and generation of transgenic flies
To generate QF2 lines, we used pENTR/D-TOPO vectors with various enhancer insertions (gifts from the laboratory of G. Rubin) as entry vectors for Gateway cloning into the pBPQF2Uw vector using LR Clonase II Enzyme mix (Invitrogen, 11791020). pBPQF2Uw was made using NEBuilder HiFi DNA assembly master mix (New England Biolabs) to replace the GAL4 on the pBPGAL4.2Uw-2 vector (Addgene, 26227) with QF2 from pBPGUw-HACK-QF2 (Addgene, 80276). The resulting constructs were sequence-verified and inserted into JK22C landing sites by Bestgene. pGP-5XQUAS-IVS-Syn21-jGCaMP8m-p10 was made using NEBuilder HiFi DNA assembly master mix (New England Biolabs) to replace the 20XUAS on the pGP-20XUAS-IVS-Syn21-jGCaMP8m-p10 vector (Addgene, 162387) with 5XQUAS from pQUAST (Addgene, 24349). Plasmids were injected to embryos at BestGene. Genetic labelling with these drivers is unlikely to disrupt normal development; a previous study showed that drivers with improved translation efficiency could increase GFP expression by 20-fold with no apparent effect on neuronal morphology37.
Immunostaining
The procedures used for fly dissection, brain fixation and immunostaining were described previously10. For primary antibodies, we used rat anti-DNcad (1:30, DSHB, RRID AB_528121), chicken anti-GFP (1:1,000, Aves Labs, RRID AB_10000240), rabbit anti-DsRed (1:500, Takara Bio, RRID AB_10013483) and mouse anti-rat CD2 (1:200, Bio-Rad, OX-34).
Confocal imaging
Immunostained brains were imaged using a laser-scanning confocal microscope (Zeiss LSM 780). Images of antennal lobes were taken as confocal stacks with 1-mm-thick sections. Representative single sections were shown to illustrate the arborization features of ORN axons and PN dendrites, with brightness adjustment, contrast adjustment and image cropping done in ImageJ.
Calculating the percentage of ORN axons matching with PN dendrites
PN dendritic pixels and ORN axonal pixels were defined by first smoothening the image using ‘gaussian blur’ (radius = 2 pixels) and then thresholding the image based on the algorithm ‘Otsu’ in Fiji. We found that this algorithm could efficiently separate the neurons of interest from the background. Irrelevant signals (such as the PN axons, cell bodies or autofluorescence) that still persisted after these operations were manually masked out in the analysis. A portion of ORN axons were considered as matching with PN dendrites if they had overlapping pixels on a single z-plane in the image. Note that the definition of glomerulus becomes vague as ORN axons and PN dendrites innervate more and more outside the original glomerulus.
The calculated overlap between ORN axons and PN dendrites is always lower than 100%. This is because ORN axons or PN dendrites do not occupy the entire glomerulus, for a technical reason and for a biological reason. Technically, if one examines axons and dendrites with super resolution, they should not overlap at all, because each physical space should be occupied by only one entity if the resolution is sufficiently high. In our quantifications, we used ‘gaussian blur’ to best recapitulate the adjacent areas of a single axon or dendrites that should be considered as ‘overlap’. This is an empirical parameter and would not achieve 100% overlap. Biologically, as well as ORN–PN synapses, both ORNs and PNs also form reciprocal synapses with antennal-lobe LNs. Regions with ORN–LN synapses lack PN dendrites; regions with PN–LN synapses lack ORN axons. Thus, ORN axons and PN dendrites don’t overlap in these regions.
In our analyses, we use the same parameters to quantify all genetic conditions. Thus, our conclusions about the changing of ORN–PN overlap under different genetic conditions should not be affected by these factors.
Ca2+ imaging and data analysis
Delivery of odour stimuli
Ten microlitres of PA (Thermo Fisher Scientific, 376910010) or ten microlitres of cVA (Cayman Chemical, 10010101) was applied to filter paper (Amazon, B07M6QJ2JX) inserted inside a 1-ml pipette tip. The pipette tip was left aside for at least 30 min before being positioned approximately 5 mm away from the fly antenna. Close positioning of PA and cVA is necessary because both odorants are large pheromone molecules with relatively low volatility. This method has also been used in other studies12. Other odorants, such as MP (Thermo Fisher Scientific, L05509.36), MM (Thermo Fisher Scientific, 165015000) and farnesol (Thermo Fisher Scientific, 119121000), were stored in a small glass bottle and delivered to the fly antenna through tubing, with a 10% dilution in heavy mineral oil on the day of experiments. A constant stream of charcoal-filtered air (1 l per min) was directed towards the fly, switching to odorant-containing air for 1 s as the odour stimulus before returning to the airstream. A pulse of charcoal-filtered air served as a negative control. Odorants, including the control pulse, were interleaved with at least 15-s intervals. Each odorant was delivered two to three times per recording, with the delivery sequence shuffled within each cycle. As described previously38, we glued flies to a custom stage. Dissection and imaging protocols also followed a previous study38.
Data acquisition and alignment
We used a two-photon microscope with a moveable objective (Ultima IV, Bruker). The two-photon laser (Chameleon Ultra II Ti:Sapphire, Coherent) was tuned to 925 nm in all of the imaging experiments. We used a ×16/0.8 NA objective (Nikon) for all imaging experiments. The laser intensity at the sample was 15–30 mW. A 575-nm dichroic split the emission light. A 490–560-nm bandpass filter (Chroma) was used for the green channel and a 590–650-nm bandpass filter (Chroma) was used for the red channel. We recorded all imaging data using a single z-plane, at a rate of 9–13 Hz. We perfused the brain with extracellular saline composed of 103 mM NaCl, 3 mM KCl, 5 mM N-Tris(hydroxymethyl) methyl-2-aminoethanesulfonic acid (TES), 10 mM trehalose, 10 mM glucose, 2 mM sucrose, 26 mM NaHCO3, 1 mM NaH2PO4, 1.5 mM CaCl2 and 4 mM MgCl2. All data were digitized by a Digidata 1550b digitizer (Molecular Devices) at 10 kHz, except for the two-photon images, which were acquired using PrairieView (Bruker) at varying frequencies and saved as TIFF files for later analysis. We used the frame triggers associated with our imaging frames (from Prairie View), recorded on the Digidata 1550b, to carefully align odorant delivery with Ca2+ imaging measurements.
Image registration
The image stacks were motion-corrected using non-rigid motion correction (NoRMCorre39) and then manually validated to check for motion artefacts.
Defining regions of interest
To analyse Ca2+ imaging data, we defined regions of interest (ROIs) in Fiji and Python for GCaMP signals from PN dendrites in one hemisphere, or both hemispheres when the PN dendritic signals were available. We treated the entire PN dendrites from one hemisphere as one ROI.
Calculating fluorescence intensities
We used ROIs, defined above, as the unit for calculating fluorescent intensities (see above). For each ROI, we calculated the mean pixel value at each time point and then used the method ∆F/F0 to calculate, where F0 is the mean of the lowest 5% of raw fluorescence values in a given ROI over time and ∆F is F − F0.
Courtship assay
Flies were collected shortly after eclosion. Male flies were housed individually, whereas female flies (Canton-S) were housed in groups of approximately ten. All females used as courtship targets were three-to-five-day-old virgins. All males tested in the experiments had not mated. Males were four to seven days old in Fig. 4 and Extended Data Fig. 8a–d and two days old in Extended Data Fig. 8e–i to lower the courtship baseline in males. All male flies were either w+, or w– but carried more than three mini-white markers from the transgenes they possessed. In single-pair courtship assays, two males (or one male and one female) were introduced into a custom-made courtship chamber with a diameter of 2 cm. In the courtship chain assay, five DA1-ORN→VA1v-PN males were introduced into a custom-made courtship chamber with a diameter of 5 cm. Courtship experiments were performed under low white light to reduce baseline courtship activity, because vision is well known to influence the vigour of fly courtship. Before being placed into the courtship chamber, flies were briefly grouped in a tube and anaesthetized on ice for less than 10 s. Once placed into the chamber, most flies were able to move immediately but did not fly away. Fly behaviour was recorded for more than 25 min with a video camera at 13 frames per second, and the first 25 min were quantified. In the single-pair male–male courtship assay, a control male and a rewired male were age-matched, and one of them was marked with an oil paint marker (Sharpie) on their thorax at least one day before the experiment. The paint was alternated between control and rewired males. LED lights (660 nm) were used to activate DA1-ORNs expressing csChrimson.
Statistics and reproducibility
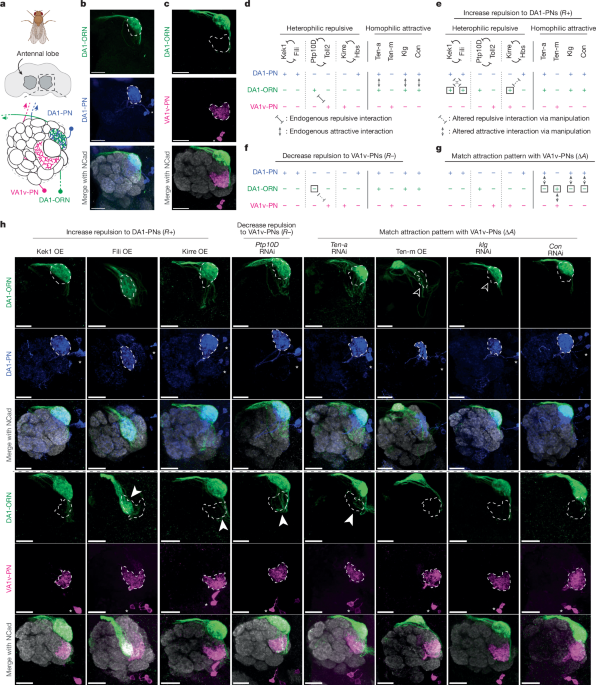
For the representative images from Fig. 1b,c and Extended Data Figs. 1a,b, 2c–f, 6c,d and 10a,b, at least five samples were examined with similar results.
Fly study design
No statistical tests were used to determine sample size. We used sample sizes (around 6–20 flies per condition) that have been shown to have sufficient statistical power in similar experiments in the past. We did not exclude flies or data from any analysis, unless brains stained for imaging appeared unsuitable (for example, broken) at the time of imaging. All experiments discussed in the paper were performed on multiple flies, with the sample size specified. For most two-photon and behavioural experiments, data across multiple days were collected with consistent results. For immunostaining, data across multiple days were collected and all imaged brains showed the same qualitative pattern of staining. Organisms are not allocated to control and experimental groups by the experimenter in this work; rather, the genotypes of the flies determine their group. Thus, randomization of individuals into treatments groups is not relevant. The investigators were not blind to fly genotype. All data collection and analysis were done computationally. During this process, data from control groups and experimental groups were analysed equally using the same well-established protocols, reducing the influence of the investigator.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.