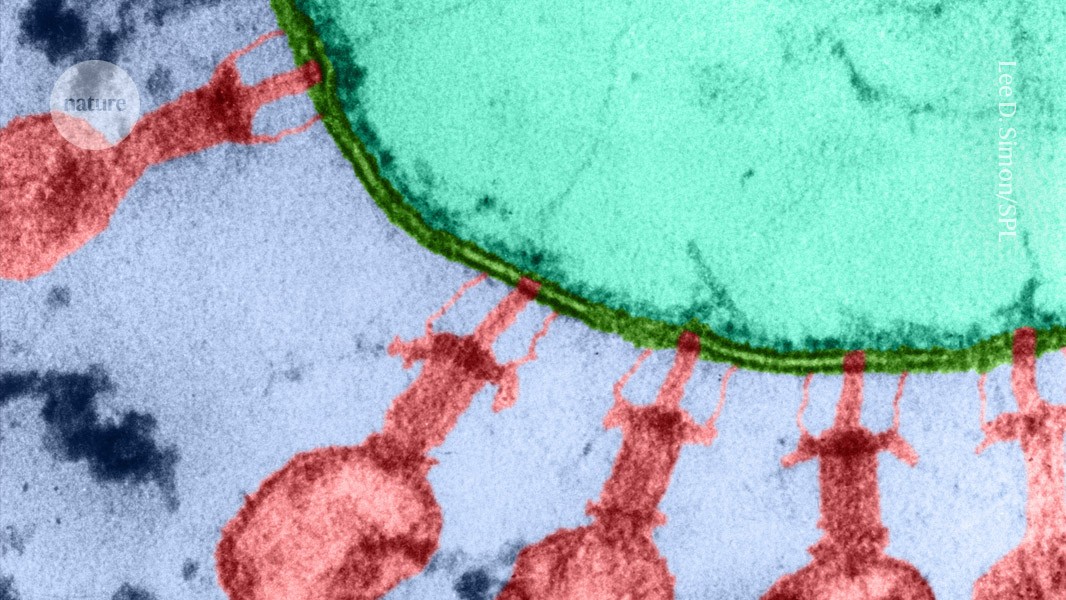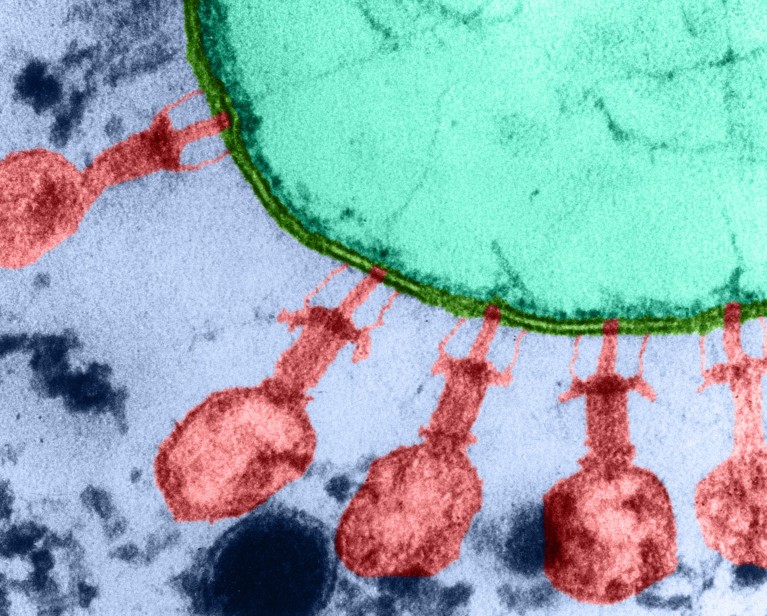
AI-designed bacteriophages were capable of infecting and killing host bacteria. Credit: Lee D. Simon/Science Photo Library
Scientists have created the first ever viruses designed by artificial intelligence (AI), and they’re capable of hunting down and killing strains of Escherichia coli (E. coli).
“This is the first time AI systems are able to write coherent genome-scale sequences”, says Brian Hie, a computational biologist at Stanford University, California. “The next step is AI-generated life”, says Hie, but his colleague Samuel King adds that “a lot of experimental advances need to occur in order to design an entire living organism”.
The study, by Hie, King and colleagues, was posted on the preprint server bioRxiv on 17 September1 and is not yet peer reviewed, but the authors say that it shows the potential of AI to design biotechnological tools and therapies for treating bacterial infections. “Hopefully, a strategy like this can complement existing phage-therapy strategies and someday augment the therapeutics [to] target pathogens of concern,” says Hie.
Genomes from the computer
AI models have already been used to generate DNA sequences, single proteins and multi-component complexes2. But designing a whole genome is much more challenging owing to complex interactions between genes and gene replication and regulation processes. These AI systems are now capable of helping scientists to manipulate highly intricate biological systems, such as whole genomes, says Hie. “There are many important biological functions that you can only access if you’re able to design complete genomes.”
To design the viral genomes, the researchers used Evo 1 and Evo 2, AI models that analyse and generate DNA, RNA and protein sequences. First, they needed a design template, which is a starting sequence that guides the AI model to generate a genome with desired characteristics. They chose ΦX174, a simple single-stranded DNA virus that contains 5,386 nucleotides in 11 genes, and all the genetic elements required to infect hosts and replicate inside them.
The Evo models had already been trained on more than 2 million phage genomes, but the researchers further trained the models — using a method called supervised learning — to generate ΦX174-like viral genomes with the specific function of infecting E.coli strains, especially those resistant to antibiotics.
Five ways science is tackling the antibiotic resistance crisis
The researchers evaluated thousands of AI-generated sequences and narrowed their search down to 302 viable bacteriophages. Most candidates shared more than 40% nucleotide identity with ΦX174, but some had completely different coding sequences. The researchers synthesized DNA from the AI-designed genomes and inserted them into host bacteria to grow phages. These phages were then experimentally tested to see whether they could infect and kill E.coli.
Some 16 of the 302 AI-designed bacteriophage showed host specificity for E. coli and could infect the bacteria. The researchers found that combinations of AI-designed phages could infect and kill three different E. coli strains, which the wild-type ΦX174 was unable to do.
“It was quite a surprising result that was really exciting for us because it shows that this method might potentially be very useful for therapeutics”, says King.