Fetal and adult tissues
Uninflamed ileum, duodenum and colon were obtained from patients undergoing tumour-resection surgery; tissues were collected at an appropriate distance from the tumour. The resection specimen was obtained as residual material after clinical procedures in accordance with the Declaration of Helsinki, the ethical guidelines of the University Medical Centre Utrecht, Utrecht, and Amsterdam University Medical Centre, Amsterdam, the Netherlands. All adult tissue material used in this study was obtained after informed consent, and with approval of tissue-specific protocols by the Medical Ethical Committee of the respective University Medical Centers.
Human fetal tissue was obtained from elective abortions at the Stichting Bloemenhove clinic in Heemstede, the Netherlands, on on the receipt of informed consent. The use of human abortion tissues was approved by the Medical Ethical Committee of the Academic Medical Center, Amsterdam, the Netherlands. Gestational age, determined by ultrasonic measurement of the diameter of the skull or femur, ranged from 19 to 21âweeks.
Generation and culturing human intestinal organoids
Human intestinal cells were isolated, processed and cultured as previously described39,40. Organoids were routinely tested for mycoplasma contamination and resulted negative. The details of organoid donors used in each experiment are listed in Supplementary Table 1.
Human intestinal organoids were split once a week by mechanic dissociation. In this study, basic culture medium includes advanced Dulbeccoâs modified Eagleâs medium/F12 (Gibco) supplemented with 100âUâmlâ1 penicillinâstreptomycin (Gibco), 10âmM HEPES (Gibco), 1Ã Glutamax (Gibco), 1Ã B-27 Supplement (Life Technologies), 1.25âmM N-acetylcysteine (Sigma-Aldrich) and 1% (v/v) recombinant Noggin (U-Protein Express).
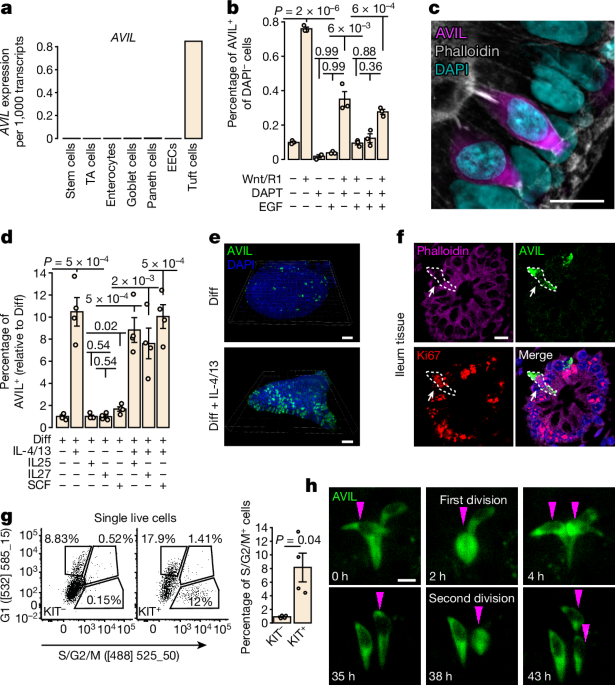
Organoids were expanded in human expansion medium as previously described39. For differentiation towards tuft cells, organoids were washed 30âmin in DMEM+++ at day 4 and the medium was replaced for tuft cell differentiation medium (diff): 0.5ânM Wnt surrogate (U-Protein Express), 20% (v/v) R-spondin1 (conditioned medium), 50ângâmlâ1 recombinant human EGF (Peprotech), 10âμM Notch inhibitor DAPT (Sigma-Aldrich) in basic culture medium. For tuft cell differentiation with IL-4 and IL-13, EGF was withdrawn, 5ângâmlâ1 human IL-4 (Peprotech) and 5ângâmlâ1 human IL-13 (Peprotech) were supplemented to tuft cell differentiation medium. BMP activation was achieved by withdrawing Noggin and addition of 50ângâmlâ1 BMP2 (Peprotech) and 50ângâmlâ1 BMP4 (Peprotech). Mature enterocytes and Paneth cell differentiation was achieved by using EGF, R-spondin1, BMP2/4 (ERB medium)41 or 10ângâmlâ1 human IL-22 (Peprotech) in WENRA42 (Wnt/R-spondin1, EGF, Noggin, ALK4,5,7 inhibitor (A83-01, Tocris)) medium, respectively. For specific experiments, 10ângâmlâ1 human IL-27 (Peprotech), 10ângâmlâ1 human IL-25 (Peprotech), 10ângâmlâ1 human SCF (Peprotech), 50ângâmlâ1 recombinant human Epiregulin (Peprotech), were used.
For organoid outgrowth experiments (Fig. 3a and Extended Data Fig. 6aâd), 100 single cells per 10âμl of basement membrane extract (BME) were seeded, and replated at day 3, keeping similar seeding density between tuft cell and non-tuft cell conditions.
For irradiation experiments, culture plates were sealed air-tight and irradiated with a single fraction of 5â6âGy (Fig. 4c and Extended Data Fig. 8jâk) using a linear accelerator (Elekta Precise Linear Accelerator 11F49, Elekta). The plates were positioned on top of 2âcm-thick polystyrene and submerged in a 37â°C water bath and radiated from below with the plate being positioned at exactly 100âcm from the radiation source.
For irradiation of AVIL lineage tracing organoids (Fig. 4gâi), organoids were differentiated for 4âdays in tuft cell differentiation medium with IL-4 and IL-13, exposed to 1âμM Tamoxifen for 20âh, then the organoids were split and irradiated (9âGy) 1âday after splitting.
Generation of stable genetically modified organoids
For reporters, for generation reporter organoids using a CRISPRâHOT approach as described in ref. 21, human duodenum, ileum and colon organoids were dissociated into small clumps, washed twice with Opti-MEM (Thermo Fisher scientific) and resuspended in BTXpress solution (BTX). Clumps were resuspended with a targeting plasmid containing a fluorescent protein (Clover, mNeon or tdTomato), which can be linearized at a defined base position by a specific single-guide RNA (sgRNA) and Cas9 (provided from a second plasmid, frame selector plasmid that also encodes mCherry). These two plasmids were co-electroporated with a plasmid encoding the sgRNA for the gene locus (Supplementary Table 2, Addgene nos. 47108, 66939, 66940, 138569, 174092). Following blasticidin selection or cell sorting based on mCherry signal, clones were picked and successful incorporation was confirmed by Sanger sequencing (Macrogen).
Regarding knock-outs, for generation knock-out organoid lines using base-editing or conventional CRISPRâCas9, spacer sequences for sgRNAs were cloned as previously described in the empty sgRNA backbone that was a kind gift from K. Joung (BPK1520, Addgene no. 65777)28. In short, plasmids were amplified using -inverse PCR (Q5, NEB), using primers with overhangs containing spacer sequences. PCR amplicons were subsequently ligated using T4 ligase (NEB) in a reaction with DpnI (NEB) to remove PCR template material. Ligations were transformed into MACH1T1 cells (Thermo Fisher) and sgRNA identity was confirmed by Sanger sequencing. For electroporation, 2.5âμg sgRNA plasmid (BPK1520, Addgene no. 65777), 7.5âμg pCMV_AncBE4max_P2A_GFP plasmid (for base-editing, Addgene no. 112100) or pCAS9-mCherry-Frame +1 plasmid (for conventional CRISPRâCas9, Addgene no. 66940), together with 10âμg PiggyBac transposon system (5âμg transposaseâ+â5âμg hygromycin resistance containing transposon)43 were co-electroporated into human duodenum, ileum and colon AVIL-Clover reporter organoids. The ATOH1â/â line was generated in human ileum wild-type organoids. After hygromycin selection, subclones were genotyped. Knock-out clones were further expanded for the following experiments. The list of guide RNAs (gRNAs) and primers to genotype can be found in Supplementary Table 2.
For FUCCI and overexpression constructs, for generation of the FUCCI reporter, ATOH1 overexpression and H2B-iRFP in human ileum AVIL-Clover reporter organoids using transposon system, constructs were cloned into a p2T-based vector44 using Gibson Assembly (NEBuilder HiFi DNA Assembly). The FUCCI sequence comprising mCherry-Cdt1-T2A-Geminin-hmAzami-Green was PCR-amplified from a construct provided as a kind gift by G. Kops (Hubrecht institute, Utrecht). The ATOH1-P2A-iRFP670 overexpression plasmid was cloned using a three-insert Gibson reaction. ATOH1 complementary DNA (IDT) was first cloned into a backbone vector with GSlinker-P2A-iRFP670, and the complete fragment of ATOH1-GSlinker-P2A-iRFP670-SV40polyA was amplified. The DNA fragment of tight TRE promoter with the ATG start codon and 3XFLAG tag was amplified from pCW-Cas9 (Addgene no. 50661). The DNA fragment of hPGK-PuroR-rTetR was amplified from pCW-Cas9. All three DNA fragments were then cloned in the digested backbone. For continuous expression H2B-iRFP670, the H2B-iRFP670 and IRES-Puromycin sequences were PCR-amplified, respectively, then were inserted into the p2T vector cut with NheI (Promega) and SmaI (Promega). Organoid lines were generated by co-electroporation of 5âµg of the respective FUCCI, ATOH1-P2A-iRFP670 or H2B-iRFP670 expression construct together with 5âµg of mT2TP transposase mediating the tol2-dependent random integration of the expression constructs into the cell genome.
For generation POU2F3 overexpression in human duodenum, colon organoids using lentivirus system, a gBlock for POU2F3 was ordered from IDT, which contained a 5Ⲡuntranslated region sequence, including an EcoRI site and a Kozak sequence, and 3Ⲡtag sequence instead of the STOP codon, which includes a Gly linker, HA tag, P2A sequence and another EcoRI site. This gBlock was cloned into a pJet vector and an EcoRI restriction enzyme cloning step was done to introduce the POU2F3 sequence into pLX vector, which enable the doxycycline-induced POU2F3-P2A-tdTomato expression31. Organoids were lentivirally transduced as previously described45.
For lineage tracing, human ileum organoids were targeted with a tdTomato and CreERT2 sequence fused with the gene encoding for Advillin using a CRISPRâHOT approach as described in ref. 21 (tdTomato-T2A-CreERT2 to the AVIL locus separated by a P2A sequence to obtain AVIL-P2A-tdTomato-T2A-CreERT2). In parallel, using mT2TP transposase-based random integration, a CAG promoter-driven loxp-flanked puromycin resistance containing a stop codon with a downstream H2B-iRFP670 was introduced (CAG-loxp-puromycin-stop-loxp-H2B-iRFP670).
Transmission electron microscopy
Organoids were chemically fixed for 3âh at room temperature with 1.5% glutaraldehyde in 0.067âM cacodylate buffered to pHâ7.4 and 1% sucrose. Samples were washed once with 0.1âM cacodylate (pHâ7.4), 1% sucrose and three times with 0.1âM cacodylate (pHâ7.4), followed by incubation in 1% osmium tetroxide and 1.5% K4Fe(CN)6 in 0.1âM sodium cacodylate (pHâ7.4) for 1âh at 4â°C. After rinsing with Milli-Q water, organoids were dehydrated at room temperature in a graded ethanol series (70, 90, up to 100%) and embedded in Epon polymerized for 48âh at 60â°C. Ultrathin sections of 60ânm were cut using a diamond knife (Diatome) on a Leica UC7 ultramicrotome, and transferred onto 50 Mesh copper grids covered with a Formvar and carbon film. Sections were poststained with uranyl acetate and lead citrate.
All transmission electron microscopy images were collected autonomously using a virtual nanoscopy slide46 on a Tecnai T12 microscope (Thermo Fisher Scientific) at 120âkV using an Eagle camera. Data were stitched, uploaded, shared and annotated using Omero v.5.6.x and PathViewer v.3.7. The final pictures were directly acquired at the microscope in a manual standard way, using the Eagle camera at 4,000âÃâ4,000.
RNA isolation and qPCR
Organoid RNA was isolated using RNAeasy kit (Qiagen), following the manufacturerâs protocol. qPCR analysis using Bio-Rad CFX Manager v.3.1 and Microsoft Excel was used to perform biological and technical replicates as previously described47. Primers are listed in Supplementary Table 3.
Flow cytometry
Organoids were dissociated into single cells using TrypLE (TrypLE Express, Life Technologies) with 10âμM Rho kinase inhibitor (Abmole) in 37â°C and mechanical disruption by pipetting every 5âmin. Cells were stained for 30âmin with antibody, then were visualized using a BD LSR Fortessa X20 4 laser (BD Biosciences, FACSDiva v.9.0) or sorted using BD FACS (fluorescence activated cell sorting) Influx (BD Biosciences, v.1.2.0.142) based on fluorescence levels. FlowJo software (v.10.6.2) was used to analyse the flow cytometry data. For scRNA-seq, single cells were sorted on FACSFusion (BD Biosciences, FACSDiva v.8.0.1) and collected in 384-well plates with ERCC spike-ins (Agilent), reverse transcription primers and deoxynucleotide triphosphates (both Promega). Single-cell sequencing was performed according to the Sort-seq method48.
For organoids staining, PE antihuman CD117 antibody (KIT; 1:100, Biolegend, 313204) or Biotin antihuman CD117 (1:100, Biolegend, 313208) and Brilliant Violet 421 Streptavidin (1:1,000, Biolegend, 405226), LIVE/DEAD Fixable Far Red Dead Cell Stain Kit (1:1,000, Thermo Fisher, L34974) were used in some experiments. For sorting from primary human fetal and adult intestine tissue, Alexa Fluor 488 antihuman CD326 (Epcam; 1:100, Biolegend, 324210), APC/Cy7 antihuman CD45 (1:100, Biolegend, 304014), PE antihuman CD117 antibody (KIT; 1:100, Biolegend, 313204) were used. For cell multiplexing oligo labelling, organoids were digested into single cells using TrypLE (Thermo Fisher), washed three times with ice-cold PBSâ+â10% FBS and incubated 15âmin at room temperature with 100âμl Cell Multiplexing Oligo. After washing, 8,000 live cells (4â²,6-diamidino-2-phenylindole negative (DAPIâ) cells) per condition were sorted into collection tube, were subjected to droplet-based scRNA-seq using the 10X Genomics platform.
Histology and immunostainings
For immunostainings, sections of formalin-fixed, paraffin embedded human colon and ileum tissue were obtained from resections performed at the University Medical Center Utrecht, the Netherlands. Anonymized archival pathology material was used according to the guidelines of the University Medical Center Utrechtâs Research Ethics Committee49. The human intestine tissues were fixed for 2âh at room temperature in 4% formalin, embedded in paraffin and stained as described previously50. Rabbit anti-Advillin (1:500, Sigma-Aldrich, HPA058864), mouse anti-Ki67 (1:4,000, monosan, MONX10283), rabbit antichromogranin A (1:600, labned.com, LN1401487) followed by goat antirabbit (ready to use, Immunologic, DPVR110HRP) or EnVision kits antimouse (ready to use, Dako, K4001) conjugated to horseradish peroxidase and then visualized (VS200 slide scanner, software v.VS200 ASW 3.3, Olympus-Lifescience).
Whole-mount staining of organoids was performed as previously described51. In brief, organoids were removed from the BME, then were fixed 2âh at room temperature in formalin. Next, the organoids were permeabilized using 0.1% Tween 20 (Sigma-Aldrich) in PBS for at least 15âmin and blocked for at least 1âh in 0.1% Triton X-100 (Sigma-Aldrich), 1âgâlâ1 bovine serum albumin (Sigma-Aldrich) in PBS. Primary antibodies used were rabbit anti-AVIL (1:300, Sigma-Aldrich, HPA058864), rabbit anti-POU2F3 (1:100, Sigma-Aldrich, HPA019652), goat anti-GFP (1:600, Abcam, ab6673), mouse anti-Ki67 (1:100, BD Pharminge, 550609), rabbit antichromogranin A (1:100, labned.com, LN1401487), rabbit antimucin 2 (1:200, Santa Cruz Biotechnology, sc-15334), rabbit anti-APOB (1:100, Novus Biologicals, NBP2-38608), rabbit anti-Lysozyme antibody (1:100, GeneTex, GTX72913), rabbit anti-Vimentin (1:100, Cell Signaling technology, 5741S), mouse anti-TM4SF4 antibody (1:100, Sigma-Aldrich, sc-293348), APC Mouse antihuman CD274 (1:100, BD Pharmingen, 563741), goat anti-GNAT3 (1:100 in organoids, LSBio, LS-B4942), goat anti-GNAT3 (1:500 in human intestine tissue slides, OAEB00418, Aviva Systems Biology), rabbit anti-CD117 (c-kit, 1:100, DAKO, A4502). Organoids were incubated with Alexa Fluor 488 phalloidin (1:1,000, Thermo Fisher Scientific, A12379) or phalloidin-atto647N (1:1,000, Sigma-Aldrich, 65906), with the corresponding secondary antibodies Alexa Fluor 405 donkey antirabbit (1:1,000, Thermo Fisher Scientific, A48258), Alexa Fluor 488 donkey antimouse (1:1,000, Thermo Fisher Scientific, A21202), Alexa Fluor 488 donkey antigoat (1:1,000, Thermo Fisher Scientific, A11055), Alexa Fluor 568 donkey antirabbit (1:1,000, Thermo Fisher Scientific A10042), Alexa Fluor 647 goat antimouse (1:1,000, Thermo Fisher Scientific, A21236), Alexa Fluor 647 donkey antirabbit (1:1,000, Thermo Fisher Scientific, A31573) and DyLight 755 donkey antirabbit (1:1,000, Molecular Probes, Thermo Fisher Scientific, SA5-10043) in blocking buffer containing DAPI (1:1,000, Invitrogen, D1306). Sections were embedded in fructoseâglycerol clearing solution, then visualized on Leica SP8 confocal microscope (LAS X v.1.1). Image analysis was performed using ImageJ (Fiji, v.1.51n) and Imaris (Andor Technology, v.x64 9.3.1) software.
Some images were obtained using a Zeiss LSM880 confocal microscope with Airyscan (Carl Zeiss, software version ZEN 2) and a LCI Plan-Neofluar Ã63 numerical aperture (NA) 1.3 water immersion objective (Carl Zeiss) at a voxel resolution of 0.04âµm (x/y) to 0.19âµm (z). Images were deconvoluted using the Zen Black (Carl Zeiss)-inbuild Airyscan postprocessing module. Images were processed (Gauss filtering) using Fiji and rendered in Imaris.
STED super resolution microscopy was performed using a Leica STELLARIS 8 STED microscope (software v.4.7.0.28176) using a HC PL APO CS2 Ã100 NA 1.40 oil objective. Organoids expressing AVIL-Clover were fixed, stained with DAPI and phalloidin-atto647N and mounted on 0.16â0.19-mm-thick cover glasses (Glaswarenfabrik Karl Hecht GmbH & Co KG) in ProLong Gold Antifade Mountant (Thermo Fisher Scientific). Tuft cell reporter and DAPI signal were recorded with confocal resolution and tuft cells identified. The actin cytoskeleton was visualized with super resolution microscopy with a pixel resolution of 10âÃâ10ânm. To do so, phalloidin-atto647N was illuminated with 647ânm excitation and 775 depletion lasers. The signal was recorded with a line averaging of 16, a dwell time of 0.69âµs.
For clustering analysis, the surface of organoids in three-dimensional image stacks was projected into two dimensions using the LocalZProjector Fiji plugin based on DAPI-stained nuclei and tuft cells were segmented using the ITK-SNAP software (v.3.8.0). For the clustering analysis, a custom-made MATLAB script was used. Inter-tuft cell distance was measured based on the Euclidean distance of their centroid position and expressed as a multiple of the average cell distance. A density map was computed, and a contour plot was generated using the imcontour function. Clusters were identified within an eight-cell-distance and analysed using the MATLAB package âDistance-based clustering of a set of xy coordinatesâ (Y. Marcon (2023), https://www.mathworks.com/matlabcentral/fileexchange/56150-distance-based-clustering-of-a-set-of-xy-coordinates), made available through the MATLAB Central File Exchange.
Live-cell imaging of human intestinal organoids
Live imaging experiments were performed on a Leica SP8 confocal laser scanning microscope equipped with Argon laser and White Light Laser at 37â°C and 5% CO2 using a Leica Ã20 NA 0.7 air objective. Images were acquired in a line-sequential mode separating the fluorophore recordings with minimal spectral overlap with a final pixel resolution of 1.65 pixels per micrometre, an axial resolution of 1.4âµm and a time interval of 9âmin.
Long-term live imaging was performed using a LS1 Live light sheet microscope (Viventis Microscopy, software v.2.0.0.3) using a Nikon Ã25 NA 1.1 water immersion objective at a magnification of Ã18. Organoids were mounted on a single-chamber sample holder 1âday before the start of imaging. A position-specific alignment of the light sheets with a thickness of 2.2âµm was done. The samples were imaged with a 488 and 561ânm illumination to visualize Clover- and tdTomato-based reporters with a time interval of 10âmin at 37â°C and 5% CO2.
The postacquisitional analysis was done with custom-made Fiji script, the H.264 encoding was done using HandBrake (v.1.8.1) software.
scRNA-seq analysis
scRNA-seq libraries of organoid-derived material, and KIT+ enriched cells from fetal and adult intestines were sequenced on an Illumina NextSeq platform, at a median sequencing depth of 49,861 reads per cell. Reads were mapped to a human genome (hg38) integrated with the Clover transcript using STAR (v.2.7.8a), reads with many mapping positions were excluded. Reads were associated with genes if they were mapped to an exon. Reads were demultiplexed and collapsed into unique molecular identifier (UMI) tables using umi_tools (v.1.1.1) allowing up to one hamming distance of the cell barcode. Cells with less than 500 UMI or with more than 40% mitochondrial genes were excluded from analysis.
All analysis was performed in R. We used the MetaCell package (v.0.3.5)26 to analyse all scRNA-seq data collected in this study. Default parameters were used unless otherwise stated. We derived a MetaCell cover of DAPIâ/AVIL-Clover+ and DAPIâ epithelial cells from human ileal organoids. Mitochondrial genes and the highly variable immunoglobulin genes (IGH, IGK and IGL prefixes) were removed from the UMI tables. Gene features for MetaCell covers were selected using the parameter Tvm=0.1, total umi more than ten and more than three UMI in at least three cells. We filtered the list of gene features used for MetaCell analysis from genes associated with cell cycle, immediate stress response and gene modules inducing strong patient-specific biases. To this end, we first identified all genes with a correlation coefficient of at least 0.13 for one of the anchor genes TOP2A, MKI67, PCNA, MCM4, UBE2C, STMN1, FOS, EGR1, IER3, FOSB, HSPA1B, HSPA1A, HSP90AA1 and DNAJB1. We then hierarchically clustered the correlation matrix between these genes (filtering genes with low coverage and computing correlation using a down-sampled UMI matrix) and selected the gene clusters that contained the above anchor genes. We thus retained 94 genes as features. We used MetaCell to build a k-nearest neighbours graph, perform boot-strapped coclustering (500 iterations; resampling 70% of the cells in each iteration) and derive a cover of the coclustering k-nearest neighbours graph (kâ=â30). Outlier cells featuring a gene expression higher than fourfold than the geometric mean in the metacells in at least one gene were discarded. Detailed annotation of the different tuft and epithelial cell subsets was performed using hierarchical clustering of the MetaCell confusion matrix. ClusterProfiler52 (v.3.14.0) and ChIPpeakAnno (v.3.20.0) were applied to perform gene functional annotation of DEGs.
scRNA-seq of passage 1âday 7 KITâ-Â and KIT+-derived organoids was performed using the Chromium Next GEM Single Cell 3â² v.3.1 platform, and sequenced on an Illumina NovaSeq6000 platform. Reads were mapped to the human genome (hg38) and demultiplexed using cellranger (v.7.1.0). Recovered cellplex barcodes were used to assign single cells to experimental batches. Single cells with less than 64 UMI of a specific cellplex barcodes were discarded from downstream analysis. Single cells with less than eightfold UMI count ratio between highest and second highest cellplex barcodes were marked as doublets and discarded from downstream analysis. Single cells with less than 1,000 genomic UMIs or more than 20% mitochondrial content failed to pass quality control and were discarded from further analysis, resulting in 10,311 quality control-positive cells.
Clustering of passage 1 KITâ-Â and KIT+-derived organoids was performed as stated above. Gene features for the MetaCell covers were selected using the parameter Tvm=0.1, total umi>15 and more than three UMI in at least three cells, resulting in 228 features.
We reanalysed scRNA-seq data from human primary intestinal tissue18. We selected 15,184 single cells from healthy adult small intestine, with more than 1,000 and less than 20,000 total UMI for further analysis. Cells were analysed with the MetaCell package as previously described to derive a two-dimensional (2D) representation of the data for Extended Data Figs. 3a and 5b. Otherwise, we used predefined annotations to epithelial cell types. In Extended Data Figs. 3j and 5f we sampled 500 cells from each cell types out of the total 77,364 healthy adult single cells in that database.
Quantification and statistics
No statistical methods were used to predetermine sample size. All experiments were performed in several distinct replicates, as indicated in the text and figure legends. Organoids analysed were chosen randomly. All statistical tests were two-tailed, except in Fig. 1b,d and Extended Data Fig. 1f, where different growth conditions were assessed for increased tuft cell numbers. We used Studentâs t-test for continuous data and MannâWhitney test for discrete data, and used FDR adjustment to correct for many hypotheses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.