Zebrafish husbandry and maintenance
Zebrafish were kept at 28 °C under a day/night cycle of 14/10 h, pH of 7–7.5, and a conductivity of 600 µS. The following transgenic lines were used in this study: Wild-type fish of the TL strain, Tg(elavl3:H2b-GCaMP6s)jf5, Tg(atf5b:QF2)mpn422, Tg(cort:QF2)mpn430, Tg(itpr1b:QF2)mpn423, Tg(sp5l:QF2)mpn421 and Tg(QUAS:GCaMP6s)mpn164. All larvae were produced by natural matings and raised until 6 or 7 days post fertilization (dpf) at 28 °C in Danieau’s solution in petri dishes. The animal experiments were performed under the regulations of the Max Planck Society and the regional government of Upper Bavaria (Regierung von Oberbayern), approved protocols: ROB-55.2-2532.Vet_03-19-86, ROB-55.2-2532.Vet_02-19-16, ROB-55.2-2532.Vet_02-21-93.
Single-cell RNA sequencing
Tissue dissection
The 6 or 7 dpf wild-type larvae were anaesthetized in tricaine (one larva at a time), followed by removal of the eye and the skin around the brain using a fine Tungsten needle. Both OT hemispheres, together with the torus longitudinalis, were carefully dissected under a stereoscope and transferred into a 1.5 ml low-binding-protein tube containing ~50 µl PBS, and placed on ice. Cell dissociation was performed immediately after the dissection. Eleven dissection sessions were performed on different days, with 20–25 OT and torus longitudinalis dissected for each batch.
Cell dissociation
Cell dissociation was performed using papain (Papain Dissociation System, Worthington Biochemical Corporation). Five hundred microlitres of oxygenated papain was added to the 1.5 ml tube containing the 50 μl PBS and the dissected tissue, and incubated at 37 °C with superficial oxygen flow. The samples were gently pipetted every 10 min, and were fully dissociated after 45 min (a 2 μl sample was visually inspected to assess cell separation). The cells were then centrifuged for 10 min at 400g, at 4 °C, using a swinging bucket centrifuge. The supernatants were removed, and the cells were resuspended in 260 μl of equilibrated ovomucoid solution and 30 μl of DNaseI. The cells were then centrifuged for 10 min at 400g, at 4 °C, the supernatants were removed, and the cells were resuspended in 500 μl PBS and 0.04% BSA. This step was repeated once again, after which the cells were filtered using 30 μm cell strainer. Finally, the cells were centrifuged, and the supernatants were removed, leaving 80 μl in which the cells were pipette gently. Five microlitres of the dissociated cells were transferred into a new 1.5 ml low-binding protein tube and mixed with 13 μl PBS, 0.04% BSA and 0.4% Trypan blue for cell counting and viability assessment.
Library preparation and sequencing
The dissociated cells were loaded onto a commercially available droplet-based single-cell barcoding system (10x Chromium Controller, 10x Genomics). The Chromium Single Cell 3′ Reagent Kit v3 (10x Genomics) was used to prepare single-cell 3′ barcoded cDNA and Illumina-ready sequencing libraries according to the manufacturer’s instructions. The cDNA libraries were sequenced using an Illumina HiSeq 2500 machine (batches 1–8) or Illumina NovaSeq 6000 (batches 9–11), with a mean of ~76,000 reads per cell over the 11 batches.
Quality check, batch correction and clustering analysis
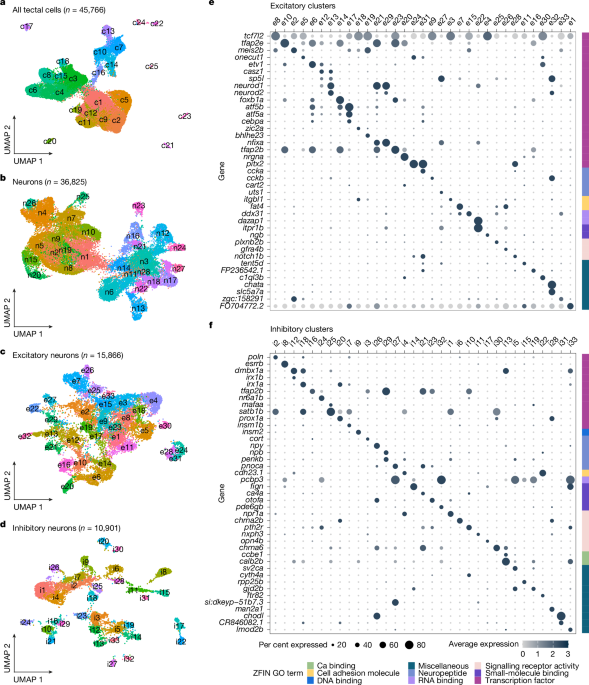
The sequenced data were processed using CellRanger-7.1.0 (filtering, barcode and unique molecular identifier (UMI) counting) with default command line options. The sequenced reads were aligned to the zebrafish GRCz11 genome assembly (Ensembl release 98). To prevent confounding effects of the tissue dissection and cell dissociation on gene expression analysis, immediate early genes (IEGs) that were induced during the procedure60 were removed prior to the analysis. We have curated a list of 43 zebrafish IEGs (Supplementary Table 2), and excluded them from the cell–gene matrix. The data from each of the 11 batches were filtered using the Seurat R package version 5.1.061, to ensure analysis of high quality cells, including filtering out cells expressing less than 600 or higher than 4,000 genes, cells with more than 7,500 UMIs, or containing higher than 10% mitochondrial genes. Additionally, to eliminate contamination from adjacent tissues, cells expressing the habenula marker genes gng8 and kiss1, as well as cells expressing the lens crystallin gene crygm2d13 above 0.5, were excluded. The data were then normalized using the Seurat LogNormalize method with a scale factor of 10,000. A set of 2,000 variable features were identified using the vst selection method, and the data were scaled using the ScaleData command. The detected variable features were used for principal component analysis. In order to filter out doublet cells from the analysis, initial clustering was performed on each batch separately, and the clustering information was used as an input for DoubletFinder62. Finally, the 11 batches were merged into a single Seurat object, and cells identified as doublets by DoubletFinder (n = 5,931) were filtered out. The resulting dataset contained 45,766 cells with a mean of 1,224 genes per cell and 2,420 UMIs. We then performed batch correction using Harmony63, grouping the variables according to their original batch followed by dimensionality reduction with UMAP64. Clustering analysis was performed by applying the Seurat FindNeighbors command with reduction=”harmony” and dims = 25 and the FindCluster command with resolution set to 0.6 (sub-clustering resolution was: neurons = 1, excitatory neurons = 1.2, inhibitory neurons = 1.2). The clustering resolutions were defined using clustering trees generated with the R package Clustree, together with SC3 stability scores65,66. Following the separation of excitatory and inhibitory cells, and prior to their respective clustering, cells expressing gad1b above 0.8 were filtered out from the excitatory dataset, while cells expressing either slc17a6a (also known as vglut2b) or slc17a6b (also known as vglut2a) above 0.8 were filtered out from the inhibitory dataset. After manual inspection of marker gene expression, clusters e24, e28 and e31 were split from a single cluster, as well as clusters i3 and i16, using the FindSubCluster command with resolutions of 0.3 and 0.25, respectively. To evaluate clustering correlation, genes were ranked according to their specificity score, calculated using the genesorteR R package67, and correlation matrices were generated according to the top ten scored genes per cluster. The vast majority of clusters showed low correlation with other clusters, representing unique t-types. Cluster robustness was evaluated by measuring pairwise Jaccard index of cells grouped together under different clustering parameters using the R package scclusteval68. Additionally, cluster stability was evaluated by bootstrapping as described in Tang et al. 202168. Eighty per cent of the cells were sampled and reclustered. The highest Jaccard index was recorded for matching clusters for each subsample. This procedure was repeated 100 times and plotted as a Jaccard Raincloud plot. In most instances, we observed that clusters with small numbers of cells were scored low, as a result of infrequent sampling of those cells.
Generally, we took a conservative approach when defining and including clusters, which were then followed by validation of marker genes in situ.
Finally, differentially expressed genes were identified using the Wilcoxon Rank Sum test integrated in the Seurat FindAllMarkers command. Pseudotime analysis was performed using Monocle369.
Multiplexed and iterative in situ hybridization chain reaction
HCR experiments were performed on Tg(elavl3:H2b-GCaMP6s)jf5 (to enable registration onto a common coordinate system) or the transgenic knock-in larvae labelling atf5b, cort, itpr1b and sp5l expressing cells. All HCR reagents including probes, hairpins and buffers were purchased from Molecular Instruments. The staining was performed according to previously published and modified protocol28,70. The larvae were anaesthetized in 1.5 mM tricaine and fixed with ice-cold 4% paraformaldehyde/Dulbecco’s PBS (DPBS) overnight at 4 °C with gentle shaking. The following day, larvae were washed 3 times for 5 min with DPBST (1× Dulbecco’s PBS + 0.1% Tween-20) to stop fixation, followed by a short 10-min treatment with ice-cold 100% methanol at −20 °C to dehydrate and permeabilize the tissue samples. Next, rehydration was performed by serial washing of 50% methanol/50% DPBST and 25% methanol/75% DPBST for 5 min each and finally 5× 5 min in DPBST. Ten to twelve larvae were transferred into a 1.5 ml tube and pre-hybridized with pre-warmed hybridization buffer for 30 min at 37 °C. Probe solution was prepared by transferring 2 pmol of each HCR probe set (2 µl of 1 µM stock) to 500 µl of hybridization buffer at 37 °C. The hybridization buffer was replaced with probe solution, and the samples were incubated for 12–16 h at 37 °C with gentle shaking. To remove excess probes, larvae were washed 4× 15 min with 500 µl pre-warmed probe wash buffer at 37 °C. Subsequently, larvae were washed 2× 5 min with 5× SSCT (5× sodium chloride sodium citrate + 0.1% Tween-20) buffer at room temperature. Next, pre-amplification was performed by incubating the samples in 500 µl of amplification buffer for 30 min at room temperature. Separately, 30 pmol of hairpin h1 and 30 pmol of hairpin h2 were prepared by snap-cooling 10 µl of 3 µM stock by incubating the hairpins in 95 °C for 90 s, and cooling down to room temperature in a dark environment. After cooling down for 30 min, hairpin solution was prepared by transferring the h1 and h2 hairpins to a 500 µl amplification buffer. The pre-amplification buffer was removed and the samples were incubated in the hairpin solution for 12–16 h in the dark at room temperature. Excess hairpins were washed the next day 3× 20 min using 5× SSCT at room temperature. Larvae were then stored long-term at 4 °C in 5× SSCT until imaging.
For iterative HCR staining, the HCR probes and hairpins were stripped using DNAseI treatment. The imaged fish were placed separately in 1.5 ml tubes and incubated in a mix of 5 µl 10x reaction buffer (Invitrogen AM2238), 5 µl Turbo DNAseI (final concentration 0.2 U µl−1, Invitrogen AM2238) and 40 µl DPBS for 4 h at 37 °C. The samples were then washed 3 × 5 min with DPBST and the complete removal of the HCR signal was validated under a confocal microscope. The HCR stripped fish were kept separated in 1.5 ml tubes and underwent another round of HCR staining and imaging, beginning from the pre-hybridization step.
HCR data were registered onto the standard brain as previously described28 using Advanced Normalization Tools (ANTs)71. For the functional HCR experiments, where iterative HCR stainings were performed, each iteration was registered to the first iteration of the same animal. A total of 3 HCR rounds were performed on single animals without any noticeable morphological distortions or reduction of the endogenous fluorescence of the Tg(elavl3:H2b-GCaMP6s) used as the reference registration channel.
HCR image registration
All HCR and immunostaining images were aligned onto the mapzebrain.org Tg(elavl3:H2b-GCaMP6s) standard brain27,28.
The ANTs registration command used was:
antsRegistration -d 3 –float 1 -o [${output1},${output2}] — interpolation WelchWindowedSinc –use-histogram-matching 0 -r [${template},${input1},1] -t rigid[0.1] -m MI[${template},${input1},1,32,Regular,0.25] -c [200×200×200×0,1e-8,10] — shrink-factors 12×8×4×2 –smoothing-sigmas 4×3×2×1vox -t Affine[0.1] -m MI[${template},${input1},1,32,Regular,0.25] -c [200×200×200×0,1e-8,10] — shrink-factors 12×8×4×2 –smoothing-sigmas 4×3×2×1 -t SyN[0.01,6,0.0] -m CC[${template},${input1},1,2] -c [200×200×200×200×10,1e-7,10] –shrink- factors 12×8×4×2×1 –smoothing-sigmas 4×3×2×1×0 followed by applying the transformation files on the HCR image channels using the ANTs command:
antsApplyTransforms -d 3 -v 0 — float -n WelchWindowedSinc -i ${input3} -r ${template} -o ${output4} -t ${output1}1Warp.nii.gz -t ${output1}0GenericAffine.mat
All the registered HCR data used in this study are publicly available through the mapzebrain.org atlas.
Confocal imaging
HCR-labelled samples were embedded in 2% low-melting agarose in 1× DPBS (Dulbecco’s PBS) and imaged using an upright Zeiss LSM700 confocal scanning microscope with a 20× water immersion objective. The images were acquired with Zeiss ZEN 2012 SP1 software (Release Version 8.1.6.484). z-stacks, comprising 2 tiles (or 1 tile for functional HCR experiment), were taken and stitched to produce a final image with size of 1,039 × 1,931 pixel (463.97 × 862.29 µm, 1 µm in z).
Pixel intensity quantification of HCR data
For each HCR-labelled gene, three larvae were imaged and registered as described above. A maximum intensity projection was generated for each image for planes z289–z291. Coronal sections were generated using the Fiji ‘reslice’ option72, and the maximum intensity projection was generated for planes x463–x465. ROIs spanning the SPV for both the transverse and coronal section were used to measure the pixel intensity profile using a custom Fiji macro. The data were analysed and plotted using R.
Tissue localization of cell bodies with HCR signal
Cell centroids in the HCR data were manually labelled using napari points layer tool73, by examination of HCR signal together with the nuclear labelling of the Tg(elavl3:H2b-GCaMP6s). For the functional imaged-HCR-labelled animals, the experimenter was blinded to the functional trace of the labelled centroids. For identifying co-labelled submarkers in atf5b– and sp5l-expressing cells, polygons were first manually drawn using napari around the cells expressing the main markers. The expression of submarkers were manually determined by labelling positive centroids within the main polygons. The centroids were then registered onto the standard brain using ANTs and the R package ANTsR. To define the molecular layers of the OT, the NNDs between centroids were measured in 3D using the R package spatstat74. The centroids were visualized using the R package Plotly, hierarchically clustered using complete linkage method implemented in the hclust function with default parameters, and plotted using the R package pheatmap75.
Functional imaging of Tg(elavl3:H2b-GCaMP6s)
Two-photon functional calcium imaging was performed on 6–8 dpf Tg(elavl3:H2b-GCaMP6s) larvae that expresses a nuclear calcium indicator in all neurons, without paralysis or anaesthesia. The animals were embedded in 2% agarose and mounted onto the stage of a modified two-photon moveable objective microscope (MOM, Sutter Instrument, with resonant-galvo scanhead) with a 20× objective (Olympus XLUMPLFLN, NA 1.0). The data were acquired with ScanImage 5.6 software. Fish that drifted along the dorsoventral axis in the preparation were excluded from analysis. Volumetric imaging of the tectum was performed with a custom-built remote focusing arm34. Refocusing through the remote arm enabled rapid sequential imaging of 6 planes (512 × 512 pixels) spanning 60–100 µm of the tectum along the dorsoventral axis at 5 volumes per second. In each fish neuronal activity in the tectum was recorded over 2× 10-min sessions to cover the whole tectum, resulting in 12 imaging planes per fish in total. Laser power out of the objective ranged from 10 mW to 15 mW.
Functional imaging of cell-type-specific transgenic lines
Animals were embedded and placed under a commercial two-photon laser scanning microscope (Femtonics) or custom-built two-photon microscope as described above34,76. Sequential single-plane imaging at either 5 fps (custom-built microscope) or between 1–1.5 fps (Femtonics) was performed at 4–10 different depths along the dorsoventral axis of the specimen for a 10-min session each, with the same stimulus set as detailed below. No anatomical stack for registration was acquired from these animals.
Visual stimulation
Visual stimuli were designed with PsychoPy and projected by an LED projector (Texas Instruments, DLP Lightcrafter 4500, with 561 nm long-pass filter) from below onto Rosco Tough Rolux 3000 diffusive film water-immersed in a 10-cm petri dish. The embedded animal was placed into a 6 cm petri dish on top of the diffusive paper in the larger dish, with a spacer in between that ensured a water film between the diffusive paper and the smaller petri dish. The fish was placed at 12 mm distance from the projection screen. The fish head was manually centred in the imaging chamber, aided by projecting crosshairs on the screen. Stimuli were shown in a fixed sequence: gratings, dot motion (8×, see below for details), gratings, OFF ramp, ON ramp, looming (2×). All stimuli consisted of black features on a red background to not interfere with the green GCaMP6s fluorescence signal. Individual stimulus presentations were separated by 20 s inter-trial intervals, except for the two loom stimuli, for which a 1-min interval was used. The whole duration of the stimulus protocol was 10 min.
Moving gratings
Gratings moving caudo-rostrally with respect to the fish were shown once at the beginning and after the dot stimuli with 20 mm width and 2 Hz temporal frequency for 20 s.
Dot motion
A black dot moving on a circular trajectory (radius, 18 mm) was shown starting in front of the fish. The dot stimulus moved either in discrete jumps at 1.5 Hz or in a perceptually smooth displacement at 60.0 Hz (projector frame rate) with the same overall speed of 5 mm s−1 (15.9° s−1), resulting in a stimulus duration of 22.6 s. Each frequency was presented using a dot diameter of 4 mm (12.7°). Both clockwise and counter-clockwise presentations were shown, resulting in four different stimuli. The whole dot stimulus set was repeated twice in each session, so eight dot stimuli were shown in succession.
OFF/ON ramp
Whole-field luminance of the projected blank image was decreased to zero over the course of 2 s, and ramped up to normal background luminance within 2 s after 20 s delay.
Looming
A linearly disk was displayed with expansion from 0.6° to 110° in 83 ms centred below the fish.
z-stack acquisition, registration and ROI matching
After each functional recording, a high-resolution 2-photon z-stack of a large subvolume of the brain, including the full midbrain region, was taken (1,024 × 1,024 pixels, 1 µm in z, 835 nm laser wavelength, plane averaging 100×). Each time series average of the 12 imaging planes were registered to this stack using the scikit-learn template matching algorithm. The 2-photon brain volume was then registered to the volume of the first round HCR confocal imaging as described in ‘HCR image registration’. To transform functional ROIs from 2-photon space into HCR space, ROI pixel coordinates were transformed first from imaging plane reference frame to 2-photon z-stack reference frame and finally to the first round HCR reference frame the by running the ANTs command antsApplyTransformsToPoints twice using the respective transformation matrices from each registration step. HCR cell centroid annotations from each round were transformed into the first round HCR reference frame, and all coordinates were used as seeds for generating 3 × 3 pixel volumes that were overlaid with registered functional ROI pixels. HCR centroids were assigned to a functional ROI based on the largest fractional overlap. Finally, all assigned as well as unassigned functional ROIs were transformed to the standard brain as described above.
Analysis of two-photon imaging data
Suite2p77 was used for motion correction, ROI detection, ROI classification, and signal extraction (time constant tau = 7 s, diameter = 4 pixels). In detail, raw volume recording files were deinterleaved into individual time series for each imaging plane. Rigid and non-rigid motion correction was performed with suite2p on a low-pass filtered time series in xy (gaussian, sigma = 4). The motion-correction shifts were applied to the raw imaging time series. ROIs were detected on fivefold downsampled and motion-corrected time series and fluorescence traces were extracted using the average pixel intensities of ROIs over time. All functional ROIs were initially thresholded based on the built-in suite2p classification algorithm iscell and an anatomical tectal mask drawn in the standard brain (mapzebrain.org).
Response score
For each stimulus, a regressor was constructed by convolving a boxcar function with an exponential decay kernel that mimics the H2B::GCaMP6s off-kinetics. The resulting 8 regressors were separately fitted to the calcium trace of each functional ROI, using a linear regression model on the stimulus time window. The response score was calculated as the product of the regression coefficient (equivalent to dF/F) and the coefficient of determination R2. For clustering and dimensionality reduction (Fig. 5), the analysis included all functional ROIs that scored above the 95th percentile of the population response score for any of the visual stimuli. Response score vectors of all high-scoring neurons excluding any t-type positive ROIs (n = 7094) were scaled to unit variance and zero mean of the overall tectal population for subsequent analysis.
Hierarchical clustering and embedding
Functional clusters were identified in a two-step analysis: First, exemplars of the 7,094 high-scoring ROIs in 8-dimensional functional space were identified using affinity propagation (sklearn.cluster.AffinityPropagation, default parameters). Exemplars with less than 10 associated ROIs were excluded from subsequent analysis. The remaining 169 exemplars were clustered using hierarchical clustering (scipy.cluster.hierarchy.linkage, method=‘complete’, metric=‘correlation’; scipy.cluster.hierarchy.fcluster, criterion=‘maxclust’). In parallel, the 2-dimensional embedding for visualizing functional space was computed from the 8-dimensional response vectors of all high-scoring ROIs using UMAP (n_neighbors=25, min_dist=0.7, metric=‘euclidean’). Functional ROIs matched with HCR labels or functional ROIs from transgenic lines were transformed into the initial UMAP space by applying the UMAP transform function to their respective response vectors.
Anatomical overlap metric
3D coordinates of each t/f-subcluster that contained more than 10 ROIs were used to generate a gaussian kernel density estimate (KDE) with scipy.stats.gaussian_kde (bandwidth = 0.75). For computing pairwise overlap, the KDEs were sampled, normalized, and the minimum of the joint KDEs was taken at each point. The overlap metric is bounded from 0 to 1 (0 if no overlap, near 1 if the same). Controls were generated by shuffling t-type labels of all ROIs in the whole dataset.
t-type classification
Response scores of t-type+ ROIs were scaled to unit variance and zero mean. An SVM classifier (sklearn.svm.SVC, kernel=‘rbf’, gamma=‘scale’, C=1) was trained on 90% of all t-type+ response vectors and gene labels. To counter the uneven distribution of t-types, the training data were upsampled to 1,000 samples per t-type. Evaluation of the classifier performance was done on 10% holdout test data. This process was repeated 20 times with permutated training/test data splits. The same classification was performed with anatomical centroid positions of t-type+ ROIs as dependent variables as well as using both response vectors and anatomical positions. Each classification was run again with shuffled marker gene labels as a negative control.
Generation of knock-in transgenic lines
The knock-in lines Tg(sp5l:QF2)mpn421, Tg(atf5b:QF2)mpn423, Tg(itpr1b:QF2)mpn424 and Tg(cort:QF2)mpn430 were generated by locus-specific insertions using CRISPR–Cas9 and the GeneWeld approach78. Guide RNA (gRNA) target sequences were identified using the CCTop tool79. The gRNA target sequences are: atf5b, 5′-ATTTGGACGTCATGCTCCAGAGG-3′; cort, 5′-GCCCCTGGAGTCCCGTCTGG-3′; itpr1b, 5′-CATCTGCTCCCTGTATGCGGAGG-3′; sp5l, 5′-AGGCTCGCAGCTCCCTTACGAGG-3′. Short homology sequences of 48 bp spanning the upstream and downstream of gRNA site were ordered as complementary oligonucleotides (MWG) and cloned into donor plasmids using the GoldenGATEway strategy80. Universal gRNAs (ugRNA)78 were introduced into the donor to release the insert from plasmid after injection. The order of components of all the donor constructs was the following: ugRNA, upstream homology arm, short GSG linker, T2A, QF2, polyA signal, downstream homology arm and second ugRNA (inverted). CRISPR–Cas9 ribonucleoprotein complex was prepared at a concentration of 1.5 μM as described before4. The gRNA was produced by annealing customized CRISPR RNA (crRNA) (IDT, Alt-R CRISPR–Cas9 crRNA) with trans-activating crRNA (tracrRNA) (IDT, 1072533) in annealing buffer (IDT, 11-05-01-12). The gRNA was incubated with Cas9 protein (IDT, 1081060) for 15 min at 37 °C and the donor plasmid was added to the injection mix, at a final concentration of 20 ng μl−1. The CRISPR–Cas9 mix was injected into Tg(QUAS:epNTR-RFP)mpn165 embryos at the single-cell stage. Positive transient expressor fish were raised and screened at adulthood for germline transmission.
Cellular tracing and morphology analysis
Single neurons were sparsely labelled either during the knock-in generation procedure in mosaic F0 animals, or by transiently microinjecting 12.5 ng μl−1 QUAS:eGFP-caax plasmid into the Tg(atf5b:QF2)mpn423, Tg(cort:QF2)mpn430, Tg(itpr1b:QF2)mpn424 or Tg(sp5l:QF2)mpn421 transgenic embryos at the single-cell stage. At 6 dpf the injected larvae were anaesthetized in a lethal dose of tricaine, fixed in 4% paraformaldehyde and immunostained with mouse anti-ERK1/2 (Cell Signaling Technology 4696S, 1:250 dilution) and Chicken anti-GFP (Invitrogen A10262, 1:250 dilution), according to a previously published protocol27.
Confocal imaging was performed as described above. Individual neurons were semi-automatically traced using the freeware NeuTube81, saved as SWC files and registered to the mapzebrain.org atlas using ANTs as described above and previously27.
The traced neurons were plotted using the R package natverse82 and manually clustered according to the projection terminals or neuropil stratification patterns. A matrix for the anatomical crossing areas for each neuron was generated with the standard brain atlas (mapzebrain.org), and the heat maps were plotted using the R package pheatmap75. Soma positions were used to generate a gaussian kernel density estimate using the R packages misc3d and oce83 and plotted using the R package rgl84.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.