NEJM CPC case reports
The case records of the Massachusetts General Hospital (MGH) are published, lightly edited transcriptions of the CPCs of the MGH (Boston, MA). In the CPC, a patient case presentation is described and then an expert physician is asked to provide a DDx and a final diagnosis, along with their diagnostic reasoning, based only on the patient’s provided medical history and preliminary test results. The published cases, organized generally as diagnostic puzzles culminating in a definitive, pathology-confirmed diagnosis, are published regularly in the NEJM. We leverage these case reports, licensed from the NEJM, to evaluate AMIE’s capability to generate a DDx alone and, separately, to aid clinicians in generation of their own differential. For this latter task, we developed a user interface for clinicians to interact with AMIE.
A set of 326 case texts from the NEJM CPC series were considered. These case reports were published over a 10-year period between 13 June 2013 and 10 August 2023. Of these, 23 (7%) were excluded on the grounds that they discussed case management and were not primarily focused on diagnosis. The articles were distributed over the years between 2013–2023 as follows—2013: n = 22; 2014: n = 34; 2015: n = 36; 2016: n = 35; 2017: n = 36; 2018: n = 16; 2020: n = 23; 2021: n = 36; 2022: n = 39; 2023: n = 26. Supplementary Table 2 contains the full list of case reports, including the title, year and issue number of each report. The 302 cases include the 70 cases used by Kanjee et al.1.
These case reports cover a range of medical specialties. The largest proportion are from internal medicine (n = 159), followed by neurology (n = 42), paediatrics (n = 33) and psychiatry (n = 10). The text corresponding to the history of the present illness (HPI) was manually extracted from each article as input to AMIE. The average (median) word count of these sections of the case reports is 1,031 words (mean: 1,044, s.d.: 296, range: 378–2,428). The average (median) character count is 6,619 characters (mean: 6,760, s.d.: 1,983, range: 2,426–15,196).
A modified version of the article, inclusive of the provided HPI, admission imaging and admission labs (if available in the case) was created for the human clinicians (see Extended Data Fig. 1). This version had redacted the final diagnosis, expert discussion of the DDx and any subsequent imaging or biopsy results (which are typical elements of the conclusion of the case challenges). Given AMIE is a text-only AI model, the admission images and lab tables were not fed into the model. However, text-based descriptions of specific lab values or imaging findings were sometimes included in the case description.
Training an LLM for DDx
Our study introduces AMIE, a model that uses a transformer architecture (PaLM 27), fine-tuned on medical domain data; alongside an interface for enabling its use as an interactive assistant for clinicians.
As with Med-PaLM 210, AMIE builds on PaLM 2, an iteration of Google’s LLM with substantial performance improvements on multiple LLM benchmark tasks. For the purposes of this analysis the large (L) PaLM 2 model was used.
AMIE was fine-tuned with long context length on a task mixture consisting of medical question answering (multiple-choice and long-form questions), medical dialogue generation and electronic health record (EHR) note summarization. The datasets used included the training splits of MultiMedQA (MedQA, MedMCQA, HealthSearchQA, LiveQA and MedicationQA)10, a proprietary dataset of medical conversations, and expert handcrafted EHR note summaries from MIMIC-III35. The capability to process long context input enables AMIE to handle tasks that require long-range reasoning and comprehension.
From MedQA (multiple-choice) we used US Medical Licensing Examination (USMLE) multiple-choice style open domain questions with four or five possible answers36. A set of 11,450 questions were used for training and 1,273 questions reserved for testing. We curated 191 MedQA questions from the training set where clinical experts crafted step-by-step reasoning leading to the correct answer. From the HealthSearchQA, LiveQA and MedicationQA datasets we used expert-crafted long-form responses to 64 questions. The medical conversations were from a dataset of 218 dialogues between a clinician and patient that are associated with a corresponding ground truth diagnosis. The dataset features respiratory (n = 168), musculoskeletal (n = 40), cardiology (n = 5), gastroenterology (n = 4) and dermatology (n = 1) dialogues. In total, 102 unique diagnoses are represented with the mean number of turns 98 (25th percentile = 85.0, 75th percentile = 113.0).
No NEJM case report data was used in the tuning process and we perform contamination experiments to assess whether performance was impacted by case text that may have been in the pretraining corpus.
Zero-shot prompting
We evaluated AMIE on each of the NEJM case studies with the following prompt: ‘You are a helpful medical assistant. You will be provided and asked about a complicated clinical case; read it carefully and then provide a diverse and thorough DDx’.
Example input
Case: [Case Text]
What are the top 10 most likely diagnoses? Be precise, listing one diagnosis per line, and try to cover many unique possibilities (at least 10). The top 10 diagnoses are:
Example output
-
1.
Sézary syndrome
-
2.
Adult T-cell leukemia/lymphoma
-
3.
Mycosis fungoides
-
4.
Atopic dermatitis
-
5.
Psoriasis
-
6.
Drug reaction with eosinophilia and systemic symptoms (DRESS)
-
7.
Graft-versus-host disease (GVHD)
-
8.
Cutaneous T-cell lymphoma, not otherwise specified
-
9.
Hypereosinophilic syndrome
-
10.
Systemic lupus erythematosus (SLE)
The AMIE user interface
The interface associated with AMIE, depicted in Extended Data Fig. 2, enables users to interact with the underlying model via text-based chat in the context of a given case description. In our study, the interface was pre-populated with a text-only representation of the HPI for a given case. Clinicians were asked to initiate the interaction by querying AMIE using a suggested prompt. Following this initial prompt and AMIE’s response, clinicians were free to query the model using any additional follow-up questions, though clinicians were cautioned to avoid asking questions about information that had not already been presented in the case. A pilot study indicated that without such a warning, clinicians may ask questions about specific lab values or imaging leading to confabulations.
For a given question, the interface generated the response by querying AMIE using the following prompt template:
Read the case below and answer the question provided after the case.
Format your response in markdown syntax to create paragraphs and bullet points. Use ‘<br><br>’ to start a new paragraph. Each paragraph should be 100 words or less. Use bullet points to list multiple options. Use ‘<br>*’ to start a new bullet point. Emphasize important phrases like headlines. Use ‘**’ right before and right after a phrase to emphasize it. There must be NO space in between ‘**’ and the phrase you try to emphasize.
Case:[Case Text]
Question (suggested initial question is ‘What are the top 10 most likely diagnoses and why (be precise)?’): [Question]
Answer:
Experimental design
In order to comparatively evaluate AMIE’s ability to generate a DDx alone and aid clinicians with their DDx generation we designed a two-stage reader study illustrated in Extended Data Fig. 3. Our study was designed to evaluate the assistive effect of AMIE for generalist clinicians (not specialists) who only have access to the case presentation and not the full case information (which would include the expert commentary on the DDx). The first stage of the study had a counterbalanced design with two conditions. Clinicians generated DDx lists first without assistance and then a second time with assistance, where the type of assistance varied by condition.
Stage 1: Clinicians generate DDx with and without assistance
Twenty U.S. board-certified internal medicine physicians (median years of experience: 9, mean: 11.5, s.d.: 7.24, range: 3–32) viewed the redacted case report, with access to the case presentation and associated figures and tables. They did this task in one of two conditions, based on random assignment.
Condition I: Search. The clinicians were first instructed to provide a list of up to ten diagnoses, with a minimum of three, based solely on review of the case presentation without using any reference materials (for example, books) or tools (for example, internet search). Following this, the clinicians were instructed to use internet search or other resources as desired (but not given access to AMIE) and asked to re-perform their DDx.
Condition II: AMIE. As with condition I, the clinicians were first instructed to provide a list of up to ten diagnoses, with a minimum of three, based solely on review of the case presentation without using any reference materials (for example, books) or tools (for example, internet search). Following this the clinicians were given access to AMIE and asked to re-perform their DDx. In addition to AMIE, clinicians could choose to use internet search or other resources if they wished.
For the assignment process, we formed ten pairs of two clinicians each, grouping clinicians with similar years of post-residency experience together. The set of all cases was then randomly split into ten partitions, and each clinician pair was assigned to one of the ten case partitions. Within each partition, each case was completed once in condition I by one of the two clinicians, and once in condition II by the other clinician. For each case, the assignment of which clinician among the pair was exposed to which of the two experimental conditions was randomized. Pairing clinicians with similar post-residency experience to complete the same case served to reduce variability between the two distinct experimental conditions.
Stage 2. Specialists with full case information extract gold DDx and evaluate Stage 1 DDx
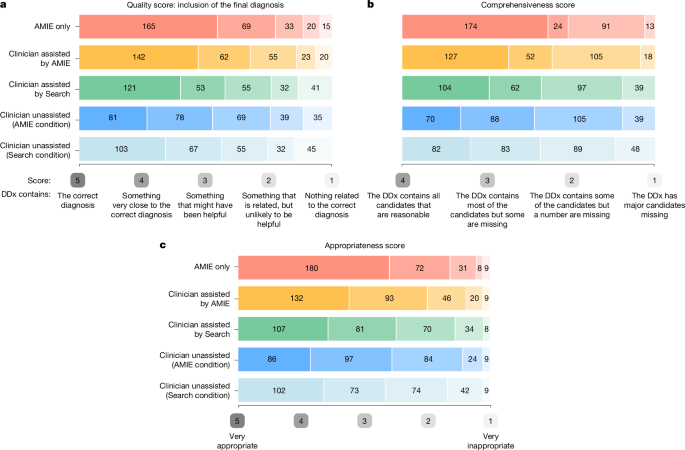
Nineteen U.S. board-certified specialist clinicians (median years of experience: 14, mean: 13.7, s.d.: 7.82, range: 4–38) were recruited from internal medicine (n = 10), neurology (n = 3), paediatrics (n = 2), psychiatry (n = 1), dermatology (n = 1), obstetrics (n = 1), and emergency medicine (n = 1). Their mean years of experience was 13.7 (s.d.: 7.82, range: 4–38). These specialists were aligned with the specialty of the respective CPC case, viewed the full case report and were asked to list at least five and up to ten differential diagnoses. Following this, they were asked to evaluate the five DDx lists generated in stage 1, including two DDx lists from condition 1 (DDx without assistance and DDx with Search assistance), two DDx lists from condition 2 (DDx without assistance and DDx with AMIE assistance) and the standalone AMIE DDx list. One specialist reviewed each case.
The specialists answered the following questions to evaluate the DDx lists:
The quality score developed by Bond et al.15 and used by Kanjee et al.1 is a differential score based on an ordinal five-point scale: ‘How close did the differential diagnoses (DDx) come to including the final diagnosis?’ The options were: 5, DDx includes the correct diagnosis; 4, DDx contains something that is very close, but not an exact match to the correct diagnosis; 3, DDx contains something that is closely related and might have been helpful in determining the correct diagnosis; 2, DDx contains something that is related, but unlikely to be helpful in determining the correct diagnosis; and 1, nothing in the DDx is related to the correct diagnosis.
An appropriateness score: ‘How appropriate was each of the differential diagnosis lists from the different medical experts compared the differential list that you just produced?’ The options to respond were on a Likert scale of 5 (very appropriate) to 1 (very inappropriate).
A comprehensiveness score: ‘Using your differential diagnosis list as a benchmark/gold standard, how comprehensive are the differential lists from each of the experts?’ The options to respond were: 4, the DDx contains all candidates that are reasonable; 3, the DDx contains most of the candidates but some are missing; 2, the DDx contains some of the candidates but a number are missing;’ and 1, the DDx has major candidates missing.
Finally, specialists were asked to specify in which position of the DDx list the correct diagnosis was matched, in case it was included in the DDx at all.
Clinician incentives
Clinicians were recruited and remunerated by vendor companies at market rates based on speciality, without specific incentives such as diagnostic accuracy or other factors.
Automated evaluation
In addition to comparing against ground truth diagnosis and expert evaluation from clinicians, we also created an automated evaluation of the performance of the five DDxs using a language model-based metric. Such automated metrics are useful as human evaluation is time and cost-prohibitive for many experiments. We first extracted the (up to ten) individual diagnoses listed in each DDx. We leveraged minor text-processing steps via regular expressions to separate the outputs by newlines and strip any numbering before the diagnoses. Then we asked a medically fine-tuned language model, Med-PaLM 210, whether or not each of these diagnoses was the same as the ground truth diagnosis using the following prompt:
Is our predicted diagnosis correct (y/n)? Predicted diagnosis: [diagnosis], True diagnosis: [label]
Answer [y/n].
A diagnosis was marked as correct if the language model output ‘y’.
We computed Cohen’s kappa as a measure of agreement between human raters and automated evaluation with respect to the binary decision of whether a given diagnosis—that is, an individual item from a proposed DDx list—matched the correct final diagnosis. Cohen’s kappa for this matching task was 0.631, indicating ‘substantial agreement’ between human raters and our automated evaluation method, per Landis & Koch37.
Qualitative interviews
Following the study we performed a semi-structured 30-min interviews with 5 of the generalist clinicians who participated in stage 1. Semi-structured interviews explored the following questions:
-
(1)
How did you find the task of generating a DDx from the case report text?
-
(2)
Think about how you used Internet search or other resources. How were these tools helpful or unhelpful?
-
(3)
Think about how you used the AMIE. How was it helpful or unhelpful?
-
(4)
Were there cases where you trusted the output of the search queries? Tell us more about the experience if so, such as types of cases, types of search results.
-
(5)
Were there cases where you trusted the output of the LLM queries? Tell us more about the experience if so, such as types of cases, types of search results.
-
(6)
Think about the reasoning provided by the LLM’s interface? Where were they helpful? Where were they unhelpful?
-
(7)
What follow-up questions did you find most helpful to ask the LLM?
-
(8)
How much time does it take to get used to the LLM? How was it intuitive? How was it unintuitive?
We conducted a thematic analysis of notes from interviews taken by researchers during the interviews, employing an inductive approach to identify patterns (themes) within the data. Initial codes were generated through a line-by-line review of the notes, with attention paid to both semantic content and latent meaning. Codes were then grouped based on conceptual similarity, and refined iteratively. To enhance the trustworthiness of the analysis, peer debriefing was conducted within the team of researchers. Through discussion and consensus, the final themes were agreed upon.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.