Plant materials and growth conditions
All the Arabidopsis transgenic lines and mutants, were in the Columbia-0 (Col-0) background except R2D2, which was in the Columbia-Utrecht (Col-utr) ecotype. The tir1-1 afb2-3 mutant15, R2D2 ratiometric reporter line16, UBQ10::IAA6-Luc18, UBQ10::IAA7-Luc, UBQ10::IAA8-Luc17 and UBQ10::IAA17-Luc19 reporters and the DR5::Luc reporter21 have all been described previously. The complementation lines pTIR1::TIR1, pTIR1::TIR1ACm1 and pTIR1::TIR1ACm3 in tir1-1 afb2-3, as well as the pTIR1::ccvTIR1, pTIR1::ccvTIR1ACm1 and pTIR1::ccvTIR1ACm3 engineering lines in tir1-1 afb2-3 were published recently4.
To generate the XVE>>axr3-mCherry transgenic line, the P88L mutation was introduced directly into AXR3(IAA17). DNA fragments encoding axr3 and mcherry with more than 15-bp overlap sequences were then assembled into the linearized pENTR/D-TOPO vector to generate the entry clone using the NEBuilder HiFi DNA Assembly kit (catalogue no. E2621L). To generate the XVE>>axr3-mCherry-KUP5, XVE>>axr3-mCherry-KUP5m, XVE>>axr3-mCherry-LRRAC1 and XVE>>axr3-mCherry-LRRAC1m in DR5::Luc background, mutations in KUP5m or LRRAC1m were introduced directly using primers. Cloned sequences were assembled with fragments encoding axr-mCherry and KUP5 or LRRAC1 together with the linearized pENTR/D-TOPO vector to generate the entry clone using the NEBuilder HiFi DNA Assembly Kit (NEB, catalogue no. E2621L). The resulting entry clone was then recombined into the destination vector pMDC7. The specific recombinant fusion sequences of XVE>>axr3-mCherry, XVE>>axr3-mCherry-KUP5, XVE>>axr3-mCherry-KUP5m, XVE>>axr3-mCherry-LRRAC1 and XVE>>axr3-mCherry-LRRAC1m are provided in Supplementary Table 1. All primers used for plasmid construction are listed in Supplementary Table 2. Final expression constructs were transformed into the Agrobacterium tumefaciens strain GV3101 by electroporation. The floral dip method was used to transform Arabidopsis plants.
Seeds were surface-sterilized using chlorine gas and sown onto half-strength Murashige and Skoog (1/2 MS) (Biochemie, catalogue no. P14170.01) medium supplemented with 1% (w/v) sucrose and 0.8% (w/v) plant agar, pH 5.9. The seeds were stratified in at 4 °C for 2 days before being grown vertically under long-day photoperiod (16 h light and 8 h dark) at 21 °C. The light sources used were Philips GreenPower light-emitting diode production modules (Philips), combining deep red (660 nm), far red (720 nm) and blue (455 nm), with a photon density of 140.4 µmol m−2 s−1 ± 3% (ref. 4).
Plant phenotypic assays
For the root formation experiment, sterilized seeds of XVE>>axr3-mCherry, XVE>>axr3-mCherry-KUP5 or XVE>>axr3-mCherry-KUP5m were sown directly on 1/2 MS medium containing 1 μM β-oestradiol or Mock and stratified at 4 °C for 2 days. Afterward, they were transferred to normal growth conditions and grown vertically for 5 days. Images were obtained using an Epson scanner (Epson Perfection V800 Photo).
For the root growth rate assay, 4-day-old seedlings were transferred onto 1/2 MS medium containing 1 µM β-oestradiol or Mock, and were then placed onto a vertical flatbed scanner (Epson Perfection V370). AutoIt scripts were used for automatic scanning at 1,200 dots per inch every 2 h as previously described27. The generated image series in ImageJ, the StackReg plugin was used for stabilization and manual tracking for root growth measurements.
For lateral root density and root growth assay, 5-day-old seedlings were transferred onto 1/2 MS medium containing 100 nM IAA or 1 µM cvxIAA or 1 µM β-oestradiol or Mock and were grown for a further 4 days. Images were obtained by Epson scanner (Epson Perfection V800 Photo). The lateral root number and main root length were quantified with ImageJ.
For root hair elongation assay, 5-day-old seedlings were transferred onto 1/2 MS solid medium containing 10 nM IAA or 100 nM cvxIAA or 1 µM β-oestradiol or Mock and grown vertically for another 1.5 or 2 days. Images were captured using a stereo microscope (Olympus, catalogue no. SZX16), and root hair length was quantified using ImageJ.
Hypocotyl segment elongation experiment was carried out as described previously27. In brief, 3-day-old etiolated seedlings were severed at the apical hook and at the shoot–root conjunction. The resulting hypocotyl segments were placed on a cellophane foil covered with depletion medium containing 10 mM KCl, 1 mM 2-(N-morpholino)ethanesulfonic acid and 1.5% phytagel, pH adjusted to 6.0 with KOH. The samples were left in the dark for 30–60 min before transfer to depletion medium supplemented with 1 µM IAA, or cvxIAA. Scanning was at 16-bit and 1,200 dots per inch every 10 min using the Autolt program. Finally, hypocotyl length was analysed with an ImageJ Macro.
Protein purification
The coding sequence of ASK1 was cloned into pGEX4T-1 vector. AFB5, IAA7 and IAA17 constructs used for protein expression in this study were published previously4. Coding domain sequences of axr2 (P88L) and axr3 (P88L) were cloned into pGEX4T-1 vector, and the resultant plasmids confirmed by sequencing were transformed into E. coli BL21 competent cells (NEB, catalogue no. C2530H) for protein purification. GST-ASK1 was obtained similarly. Expression and purification of the recombinant GST-tagged proteins from E. coli, procedure for removal of the GST tag and the use of the ÄKTA start system (GE Healthcare) were all described previously4.
Pull-down assays
In vitro pull-down assays were performed to detect the interaction of TIR1 and its mutated variants with GST-ASK1. The coding sequence of TIR1-Flag was cloned into the pF3A WG (BYDV) Vector (Promega, catalogue no. L5671). Relevant mutated variants of TIR1 were amplified directly from the previously reported plasmids4. The corresponding TIR1-Flag and its mutated proteins were obtained through in vitro translation using the TnT SP6 High-Yield Wheat Germ Protein Expression System (Promega, catalogue no. L3260). Pull-down analysis and western blotting were as previously described4. Horseradish peroxidase-conjugated FLAG monoclonal antibody (Sigma-Aldrich, catalogue no. A8592) and horseradish peroxidase-conjugated GST monoclonal antibody (Agrisera, catalogue no. AS18 4188) were diluted at 1:2,000 in 5% skim milk for western blotting.
In vitro AC activity assay and in vivo cAMP measurement
In vitro AC activity assays were performed following our previous publication4. For the in vivo cAMP measurement, 5-day-old XVE>>axr3-mCherry seedlings were first pre-treated with 20 ml of 1/2 MS liquid medium (Mock) or the same medium containing 1 μM β-oestradiol for 4 h to ensure that axr3 is sufficiently induced. Next, the pretreatment medium was replaced with Mock containing 100 nM IAA or 100 nM IAA together with 1 μM β-oestradiol. Root tissues were collected at the indicated time points and were frozen immediately in liquid nitrogen. The method for cAMP isolation and further quantification by liquid chromatography with tandem mass spectrometry has been described previously4.
Confocal microscopy of R2D2 and axr3-AC lines and quantification
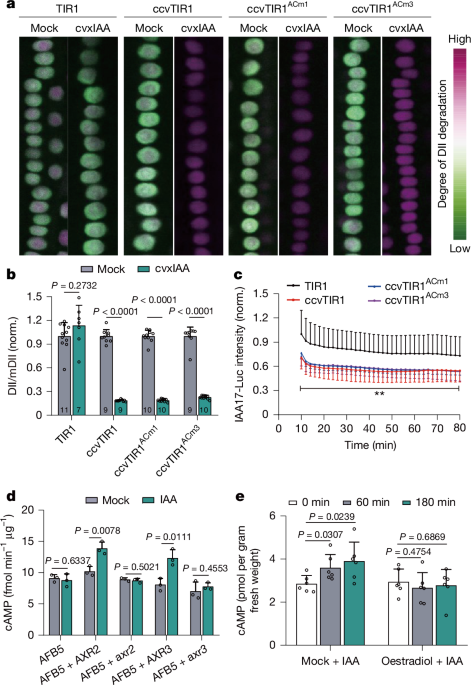
The pTIR1::TIR1, pTIR1::ccvTIR1, pTIR1::ccvTIR1ACm1 and pTIR1::ccvTIR1ACm3 homozygous transgenic lines were crossed with the R2D2 reporter line. The F1 seedlings were used directly for subsequent analysis. Briefly, 4-day-old seedlings were transferred onto solid medium with or without 200 nM cvxIAA and incubated for 1 h. Afterward, the seedlings, together with the medium, were transferred to the chamber (Thermo Fisher Scientific, catalogue no. 155361) and imaged with a vertical Zeiss LSM800 confocal microscope. All parameters were fixed during imaging to enable comparison of R2D2 signals among different genotypes. To quantify the DII-Venus and mDII-tdTomato signals, image analysis was conducted in ImageJ, and the fluorescence intensity of DII-Venus was divided by that of mDII-tdTomato to obtain the DII/mDII ratio. The whole meristem zone was selected for image analysis.
Sterilized seeds of XVE>>axr3-mCherry or XVE>>axr3-mCherry-KUP5 or XVE>>axr3-mCherry-KUP5m were germinated on 1/2 MS medium. Four-day-old seedlings were then transferred to medium with 1 µM β-oestradiol. Subsequently, the seedlings, along with the medium, were placed in a chamber (Thermo Fisher Scientific, catalogue no. 155361) and visualized using a vertical Zeiss LSM800 confocal microscope. Imaging parameters were kept consistent throughout the experiment to allow reliable comparisons of mCherry intensity across the different genotypes.
Luc imaging and quantification
Bioluminescence was imaged and quantified as described27,28. The control TIR1 (pTIR1::TIR1), pTIR1::ccvTIR1, pTIR1::ccvTIR1ACm1 and pTIR1::ccvTIR1ACm3 homozygous transgenic lines were crossed with IAA6, IAA7, IAA8 and IAA17-Luc and DR5::Luc reporter line. Four-day-old F1 seedlings were used for imaging. For steady-state analysis, the seedlings were transferred onto 1/2 MS solid Mock medium or 1/2 MS solid medium containing 200 nM cvxIAA, incubated vertically for 6 h under normal growth conditions and then used for imaging. For timecourse imaging, the seedlings were transferred to 1/2 MS solid medium containing 200 nM cvxIAA using cellophane foil. Then, 1 mM d-luciferin in 1/2 MS liquid medium containing 200 nM cvxIAA or Mock was dropped onto roots before imaging. The seedlings were then live-imaged in a Lumazone Manual Stage Dark Box (Photometric, catalogue no. LMZ-DRK-BOX) and luminescence was captured using a Photometrics Evolve 512 EMCCD camera equipped with a 17-mm fixed lens/0.95 and a second125-mm lens. The multiplier CMCCD gain was set to 300 and 150 for IAA17-Luc and DR5::Luc, respectively. The exposure time was 90 s and the time interval was 2 min.
For DR5::Luc imaging in XVE>>axr3-mCherry-KUP5 or -KUP5m and XVE>>axr3-mCherry-LRRAC1 or -LRRAC1m background, 4-day-old seedlings were transferred to 1/2 MS medium containing 1 μM β-oestradiol, and 1 mM d-luciferin was applied. Images were taken in the Lumazone Manual Stage Dark Box as described above. The multiplier CMCCD gain was set to 900, the exposure time was 90 s and the time interval was 2 min. Images were analysed in ImageJ, and the whole signal areas of similar width were selected for each root to quantify signal intensity27,28.
Transcriptome sequencing and RT–qPCR
Five-day-old seedlings were transferred into 1/2 MS liquid medium (Mock) or medium containing 200 nM cvxIAA for 3 h. Whole seedlings were collected for RNA extraction, RNA-seq and RT–qPCR analysis. All treatments were conducted with three biological replicates. Transcriptome sequencing services were provided by LEXOGEN (https://www.lexogen.com/). Differential expression analysis use the DESeq2 package29. Expression amounts were quantified as fragments per kilobase of transcript per million mapped reads. All genes showing an upregulation with a log2 fold change ≥ 1 after 200 nM cvxIAA treatment were selected for heatmap generation using the OmicsStudio platform (https://www.omicstudio.cn/tool/4). The RNA-seq datasets are available from the National Center for Biotechnology Information, under SRA accession numbers SAMN43777066–SAMN43777083. The treatment condition for RT–qPCR in the XVE>>axr3-mCherry transgenic line was consistent with that described for in vivo cAMP measurement. For RT–qPCR assay, RNA extraction was carried out using the RNeasy Plant Mini Kit (Qiagen, catalogue no. 74904). Following the manufacturer’s instructions of the RevertAid First Strand cDNA Synthesis Kit (Thermo, catalogue no. K1622), genomic DNA was removed, and 1 μg of total RNA was used for reverse transcription. The cDNA was diluted 20-fold before qPCR. Samples were prepared in three technical replicates using the Automated workstation Biomek i5 (Beckman Coulter). qPCR was performed using the LightCycler 480 (Roche) with Luna Universal rtPCR Master Mix (NEB, catalogue no. M3003S). The sequences of gene-specific primers used are listed in Supplementary Table 2. Relative gene expression was calculated using the ΔΔCT method with PROTEIN PHOSPHATASE 2A SUBUNIT A3 (PP2A) as the reference gene.
Software and statistical analysis
All graphs were generated using GraphPad Prism v.8.0.1. Figures were prepared with Adobe Illustrator 2021. One-way ANOVA, two-way ANOVA, t-test and multiple comparisons were performed using GraphPad Prism v.10.2.2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.