Patients and sample acquisition
Peripheral blood and bone marrow samples were obtained from patients with chronic phase CML. We selected patients who responded to first-line TKI (PD51633, PD51632, PD51635 and PD57332), those that did not respond to first-line TKI (PD57334, PD57335 and PD57333) and those with slow response (PD56961 and PD51634). Patients with advanced-phase CML were also included. Patients provided informed, written consent for the use of their samples for research. Patients were selected to include a wide range of ages at diagnosis and variable treatment outcomes. The study was covered under NHS Research Ethics Committee approval numbers 05/MRE/44 and 18/EE/0199.
In vitro expansion of single-cell-derived blood colonies
MNCs were isolated from peripheral blood or bone marrow samples using Lymphoprep TM (Stem Cell Technologies). Single-cell suspensions of MNCs were grown in semisolid methylcellulose-based medium MethoCultTM H4034 Optimum (Stem Cell Technologies) for 10–14 days as previously described7. Colonies were individually picked and lysed in 45 μl of RLT buffer (Qiagen).
RT–PCR for BCR::ABL1 transcript
Transcript types from patients PD56961, PD57332 and PD57335 were determined using a standardized PCR with reverse transcription (RT–PCR) protocol46. Primers used were BCR-e1-A: GACTGCAGCTCCAATGAGAAC (BCR exon 1), BCR-b1-A: GAAGTGTTTCAGAAGCTTCTCC (BCR exon 12–13) and ABL-a3-B: GTTTGGGCTTCACACCATTCC (ABL1 exon 3). This yielded a 344-bp PCR product for transcript e13a2 (6 bp (e12) + 106 bp (e13) + 175 bp (a2) + 57 bp (a3)) and a 419 bp PCR product for transcript e14a2 (6 bp (e12) + 106 (e13) + 75 (e14)+ 175 (a2) + 57 (a3)).
DNA library preparation, sequencing and read alignment
10–20 μl of lysed colony suspensions underwent WGS library preparation using the ‘laser capture micro-dissected biopsy’ pipeline47 with eight cycles of PCR. This pipeline enables the generation of high complexity WGS libraries from an input of 150–200 cells. Samples with more than 2 ng μl−1 of generated library DNA were used for paired-end, 150-bp reads WGS with Illumina NovaSeq 6000 machines. Reads were aligned to the human reference genome (GRCh38, NCBI) using the BWA-MEM (Burrows–Wheeler Aligner) algorithm.
Somatic mutation identification and filtering
SNV were identified using CaVEMan48 for each colony by comparison to an in silico unmatched sample (PD38Is_wgs). CaVEMan was run with the ‘normal contamination of tumour’ parameter set to zero, and the tumour or normal copy numbers set to five or two. Reads supporting an SNV had to have a median BWA-MEM alignment score greater than or equal to 140 and median number of soft clipped bases of 0. Further filtering designed for this bespoke pipeline was applied (https://github.com/MathijsSanders/SangerLCMFiltering). The use of the unmatched normal meant that this process called both somatic and germline SNVs. The removal of germline SNVs and artefacts of sequencing required further filtering. As published in ref. 7, we used pooled information across colonies and read counts from a matched germline WGS buccal sample to ensure that genuine somatic variants that may be present in the germline sample, either as embryonic variants or due to tumour-in-normal contamination, were also identified. Short insertions and deletions (indels) were called using cgpPindel49 with the standard WGS cgpPindel VCF filters applied, except the F018 Pindel filter was disabled as it excludes loci of depth of less than ten. Copy number aberrations (CNA) were identified using ASCAT50 by comparison to a matched normal sample or a wild-type colony from the same individual. The union of colony SNVs and indels was then taken, and reads counted across all samples belonging to the individual (colonies and buccal samples) using VAFCorrect. This allows mutations detected in any one or more colonies to be identified across all other colonies to fully capture the pattern of sharing of mutations across the colonies from an individual patient. This generates a data matrix of the number of reads supporting every mutation, depth of the sequencing of that site, and the variant allele fraction across every colony for a patient. Brass and GRIDSS pipeline51 was used for calling structural variants. Both cgpPindel and Brass used PDv38is_wgs as an unmatched normal sample.
Creating a genotype matrix
The genotype at every locus within each sample was 1 (present), 0 (absent) or NA (unknown). We inferred the genotype in a depth sensitive manner. We assumed the observed mutant read count for a colony at a given site was MTR ~ binomial(n = depth, P = expected VAF), if the site was mutant, and MTR ~ binomial(n = depth, P = 0.01), if the site was wild-type. The genotype was set to the most likely of the two possible states provided one of the states was at least 20 times more likely than the other. Otherwise, the genotype was set to missing (NA). The expected VAF was 0.5 for autosomal sites, but for chromosome X, Y and CNA sites, it was set to 1/ploidy. For loss-of-heterozygosity sites, genotype was overridden and set to missing if it was originally 0.
Phylogenetic tree topology
We constructed phylogenetic tree topologies using maximum parsimony with MPBoot52. This method minimizes the number of changes required to reach the set of mutations assigned to each sample. The inputs for MPBoot were the binary genotype matrices with missing values per individual. These were exported as a multiple alignment fasta file with one line per colony with mutant represented as A, wild-type as T and missing as ?. The command line used was: mpboot -f <fasta>-bb 1,000.
Donor with no wild-type colonies
No wild-type colonies were captured for PD57332. The tree was constructed as above, which resulted in just the mutant clade being present. Approximate features of the mutant ‘truncal’ branch were inferred as follows: (1) estimating the duration of the mutant clade as the average root to tip SNV burden divided by the cohort average BCR::ABL1 SNV mutation rate, (2) the duration of the trunk was inferred (age at sample-height of the mutant clade) and (3) the length of the trunk in molecular time was then taken as the (duration of trunk) × (cohort wild-type SNV mutation rate) + (expected mutation burden at birth).
Driver annotation
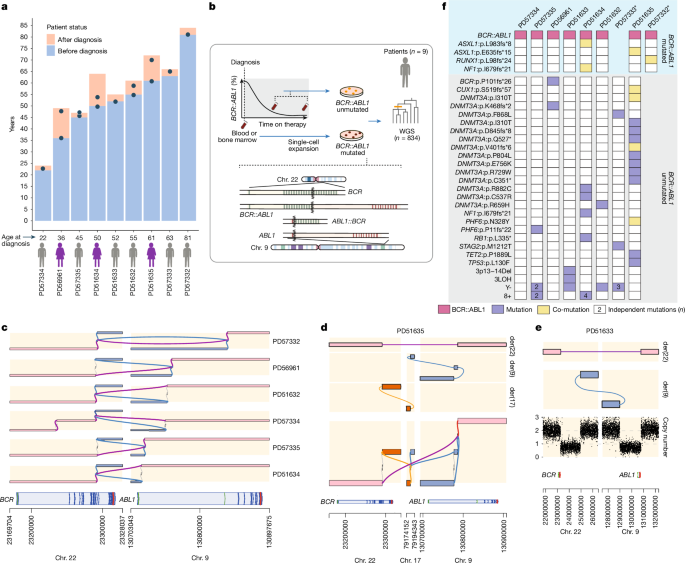
As each branch on a phylogenetic tree was assigned with SNVs and indels, it was possible to screen all branches for the presence of potential driver mutations. For this purpose, a previously7 composed list of genes (n = 35), common in clonal haematopoiesis and myeloproliferative disorders, was used: ASXL1, BCOR, CALR, CBL, CSF3R, CUX1, DNMT3A, EZH2, GATA2, GNAS, GNB1, IDH1, IDH2, JAK2, KIT, KRAS, MLL3, MPL, NF1, NFE2, NRAS, PHF6, PPM1D, PTPN11, RB1, RUNX1, SETBP1, SF3B1, SRSF2, SH2B3, STAG2, TET2, TP53, U2AF1 and ZRSR2, with the addition of ABL1 and BCR, totalling 37 genes. Branches with identified drivers were highlighted in colour on the phylogenetic trees. Annotation for BCR::ABL1 fusion was added manually to the branch on the basis of its presence or absence in individual colonies from Brass and GRIDSS results.
Timing branches
Given the linear accumulation of somatic mutations with age, we can infer the time point in life when driver mutations in phylogenetic trees had occurred. Branches at the top of a tree comprise mutations acquired at a young age, with branches lower down representing mutations arising later in life. We used our previously developed method rtreefit (https://github.com/nangalialab/rtreefit) for converting trees in which branch lengths are expressed in molecular time (that is, number of mutations) into trees in which the branch lengths are expressed in units of time (years)7. In brief, the method jointly fits separate wild-type and mutant constant mutation rates (that is, number of SNVs accumulated per year) and absolute time branch lengths using a Bayesian per individual tree-based model under the assumption that the number of observed mutations assigned to a branch is Poisson distributed with mean = branch duration × sensitivity × mutation rate, and subject to the constraint that the root to tip duration is equal to the age at sampling. Furthermore, the method accounts for an elevated mutation rate during embryogenesis by assuming an excess mutation rate through development. In running rtreefit, the mutant clade was defined as not including the trunk, so the method assumed that BCR–ABL1 was acquired at the end of the trunk. The donor PD57332 had no wild-type samples so an in silico wild-type outgroup with a long branch (equal to 1,000 × cohort wild-type mutation rate) was added. This enabled PD57332 to be processed by rtreefit in a similar way to the other donors. The rtreefit algorithm was run with four chains and 20,000 iterations per chain. Mutations were assigned to the tree in a depth sensitive manner using treemut with mutations being hard-assigned to the highest probability branch (https://github.com/nangalialab/treemut). Branch lengths were adjusted for the branch-specific SNV detection sensitivity7, in which the sensitivity of detection of fully clonal SNV variants was directly estimated from the per colony sensitivity for detecting germline heterozygous SNVs together with a multiplicative correction for the clonality (VAF) of the colonies. In calculating mutation burden and branch lengths, CNAs present in any colony in an individual were uniformly masked in all colonies for that individual and then the overall mutation burden was scaled back up by the reciprocal of 1 − expected number of mutations in the masked region. In addition to SNVs, indels and the BCR–ABL1 fusion, colonies showed a variety of CNA events. These events were curated as being present or absent in each of the colonies giving an event genotype vector like that obtained for SNVs and indels. Once the tree topology was inferred using the SNV genotypes, the branches that exactly matched the event genotype were identified and the event assigned to the corresponding branch.
Quality control of a phylogenetic tree topology
The initial quality assessment step of phylogenetic trees included the removal of colonies for which the sensitivity of CaVEMan somatic mutation detection was below 60%. Colonies that might have been contaminated with cells from another colony were also excluded. If the colony was not clonal, then the mean VAF of SNVs assigned to its private branch would be lower than 50%. To ensure that samples were clonal, colonies were excluded if the VAF of SNVs that mapped to their private branches was significantly lower than 0.4. If the VAF of mutations assigned to a colony was not 0 in non-ancestral branches, then that colony was also removed because of this indicating that cells from this colony were contaminating other colonies.
Per sample BCR::ABL1 mutation status
To ascertain BCR::ABL1 mutation status for each sample, we undertook a ‘joint’ genotyping methodology using GRIDSS. For each patient, Brass structural variant calls were reviewed for all events of interest pertaining to the BCR::ABL1 fusion event that included a core ‘target’ region consisting of the gene regions of ABL1 and BCR. For PD51635, we added RBFOX3 (chromosome 17), and for PD51633, we incorporated the large deletions present on der(9). For each patient, all samples were analysed together over the target regions to jointly identify evidence for structural variants. Per-patient results were reviewed and visualized with gGnome (https://github.com/mskilab-org/gGnome), which allowed the reconstruction of derivative chromosomes in patients with complex events such as PD51635. To ascertain the consequence of translocations, GRASS (Gene Rearrangement Analysis System, https://github.com/cancerit/grass) was used. In PD51635, a complex structural variant t(9;17) event (inversion ‘chr. 9 130776970GG]chr9:130786796]’ and translocation ‘chr. 9 130777016T [chr17:79184121[ATT’) was simplified to an equivalent single translocation event (chr. 9: +:130777016, chr. 17: +:79184121, AT) for annotation (Fig. 1d and Supplementary Table 2).
Prediction of consequence of exonic BCR::ABL1 breakpoints
In three patients (PD56961, PD57332 and PD57335) we identified breakpoints in BCR exons (exon 14, 15 and 15, respectively). To identify stop codons, we continued the reading frame from the breakpoint, adjusting for any inserted bases within the translocation event. We used SpliceAI53 to predict the splicing probabilities for each respective fusion sequence. Fusion sequences were reconstructed using GRCh38 reference sequence for BCR (chr. 22: 23170509–23328037) and ABL1 (chr. 9: 130825254–130897675) incorporating a 10-kb flanking sequence and any extra bases from the BCR::ABL1 translocation event (Extended Data Fig. 2a). Reconstructed fusion and reference sequences were used as input in the ‘custom sequence’ script (https://github.com/Illumina/SpliceAI), and ‘raw’ splice acceptor and donor probabilities from SpliceAI were converted to bedGraph format and reviewed on IGV54.
Mutational signature analysis
De novo mutation signature extraction was performed using HDP (https://github.com/nicolaroberts/hdp) with the mutations assigned to individual branches being treated as samples. Branches with fewer than 50 mutations were grouped into two per donor groups; those short branches that end before 100 mutations molecular time were pooled in the ‘early life’ group and the rest of the short branches were into the ‘late life’ group. These groups were also treated as samples. HDP extraction was then run across four chains with the following parameters for hdp_posterior: burnin=10,000, n=500, spacing=250. SBSblood signature55 was downloaded and collated with the Pan-Cancer Analysis of Whole Genomes signatures (https://cog.sanger.ac.uk/cosmic-signatures-production/documents/COSMIC_v3.3.1_SBS_GRCh38.txt). De novo extraction identified three signatures SBSblood, SBS1 and SBS18, which showed the following cosine similarities to their respective published Pan-Cancer Analysis of Whole Genomes and/or SBSblood signatures: 0.927, 0.942 and 0.884. We refitted per-donor branch groupings and individual branches against the above published versions of SBSblood, SBS1 and SBS18. This signature attribution was carried out for each of these per-donor categories or branches using sigfit::fit_to_signature with the default ‘multinomial’ model. The per-branch attributions were then carried out by (1) assigning a per-mutation signature membership probability and then (2) summing these signature membership probabilities over all SNVs assigned to a branch to obtain a branch-level signature attribution proportion. The per-mutation signature membership probability was calculated using:
$$P(\rmm\rmu\rmt\rma\rmt\rmi\rmo\rmn\,\epsilon \,\rmS\rmi\rmg)=\frac\rmS\rmi\rmg)P_(\rmS\rmi\rmg)\rmS\rmi\rmg^\prime )P_(\rmS\rmi\rmg^\prime )$$
where the prior probability, P0(Sig), is given by the mean Sigfit attribution probability of the specified signature, Sig, for the category that the mutation belongs to. The cohort level analysis of mutational signatures and C>T at CpG representation for branch categories was carried out using a random effects meta-analysis using the rma function in the ‘metafor’ R package.
Growth rate estimation
The growth rate of BCR–ABL1 clones was estimated using the previously described phylofit approach7,9. In brief, phylofit is a Bayesian approach that estimates the growth rate by directly fitting a three-parameter logistic growth curve trajectory using the joint probability density of coalescence times given the population size trajectory. The three parameters estimated by this method are the saturation population size N, the exponential phase growth rate s and the midpoint of the curve t(m). Given that patients with CML present with a high BCR::ABL1 fraction we set the upper bound of the prior for the midpoint to be the age at diagnosis and the lower bound to be the age of the MRCA of the mutant clade. We adopted uniform priors on the growth rate, s, and on the log scale saturation population size. Now, the annualized growth rate is given by S = exp(s) − 1 and the uniform priors for s, midpoint t(m) and population size N, are:
$$\beginarrayc\,\,\,\,s \sim \rmU\rmn\rmi\rmf\rmo\rmr\rmm(0.001,30)\\ \,\,\,t^(m) \sim \rmU\rmn\rmi\rmf\rmo\rmr\rmm(\rma\rmg\rme\,\rmo\rmf\,\rmM\rmR\rmC\rmA,\rma\rmg\rme\,\rma\rmt\,\rmd\rmi\rma\rmg\rmn\rmo\rms\rmi\rms)\\ \,\log _10(N) \sim \rmU\rmn\rmi\rmf\rmo\rmr\rmm(4,7)\endarray$$
The choice of upper bound for t(m) was motivated by the observation that patients with CML generally show a high BCR–ABL1 burden at diagnosis. The input data for phylofit was the time-based trees obtained using rtreefit as described above. Note that the branch lengths of the input trees were chosen to be the mean of the posterior branch lengths. The trees were restricted to the earliest sampling point. These time points are all at diagnosis or after diagnosis. Comparison of estimates were checked using an alternative recently published method cloneRate24 as detailed in Supplementary Note 3.
Telomere analysis
Telomerecat (v.4.0.2, https://github.com/cancerit/telomerecat) was used to estimate mean telomere length (bp) with (-t 75) to ameliorate the impact of NovaSeq sequencing artefacts (further details in Supplementary Note 2).
Mixed models
Linear mixed models used for SNV burden and telomere analysis were implemented in the R package lme4 to estimate the impact of age and mutant status. Age (age_at_sample_exact) was defined as the count of completed years from birth at sampling and sample mutant status (BCR_ABL1) was defined as BCR::ABL1 positive (Mt) or negative (Wt). For each response variable (y), we first tested the significance of mutant status (model 0 versus model 1) and then further terms were added to see whether this improved the model compared to the base model (model 1).
$$\beginarrayl\rmmodel\_0=y \sim 1+\rmage\_\rmat\_\rmsample\_\rmexact+(1| \rmpatient)\\ \rmmodel\_1=y \sim 1+\rmage\_\rmat\_\rmsample\_\rmexact+BCR\_ABL1+(1| \rmpatient)\\ \rmmodel\_2=y \sim 1+\rmage\_\rmat\_\rmsample\_\rmexact+BCR\_ABL1\\ \,\,\,\,\,+(1+\rmage\_\rmat\_\rmsample\_\rmexact| \rmpatient)\\ \rmmodel\_3=y \sim 1+\rmage\_\rmat\_\rmsample\_\rmexact+BCR\_\rmABL1\\ \,\,\,\,\,+(1+BCR\_ABL1| \rmpatient)\endarray$$
Models were fitted with default lme4 parameters. If a model did not converge, lme4::allFit() was used to refit the model to all available optimizers (provide by the lme4, optimx and dfoptim R packages), the best optimizer was selected from non-singular and converged refits with the highest negative log-likelihood. Only non-singular and converged models were considered for model selection using the Bayesian information criterion (BIC). For the final selected model, 95% confidence intervals (percentile bootstrap intervals) were calculated for each fixed effect, using confint(type=‘perc’) from the first 1,000 converged and non-singular parametric bootstrapped models generated using the bootstrap() function from the R package lmeresampler, using a seed (1234) for reproducibility.
SNV burden models
We first removed all samples (n = 35) from PD57332 as we were only able to grow BCR::ABL1 colonies and therefore our estimations of SNV mutation burden (nsub_adj) were expected to be biased; this left 799 samples across eight patients. The final model used is shown below and was found to have the lowest BIC value (8,645.52), as detailed in Supplementary Note 1.
$$\beginarrayl\rmSNV\;\rmmodel\,3=\rmnsub\_\rmadj \sim \rmage\_\rmat\_\rmsample\_\rmexact+BCR\_ABL1\\ \,\,\,\,\,\,+(1+BCR\_ABL1| \rmpatient)\endarray$$
Telomere length models
To account for reported issues with telomere estimation using Telomerecat and NovaSeq sequenced data9, we explored any batch effect of library preparation (library.cluster) and sequencing run (run_id.uniq) within wild-type samples (n = 469) on mean telomere length (length). The addition of sequencing run as an extra random effect (1| run_id.uniq) resulted in the lowest BIC model (7,483.33). This term was added to a series of models tested on the full dataset (n = 834), with the final model below having the lowest BIC value (13,265.15), as detailed further in Supplementary Note 2.
$$\beginarrayl\rmTelomere\;\rmmodel\,3=\rmlength \sim \rmage\_\rmat\_\rmsample\_\rmexact+BCR\_ABL1\\ +(1+BCR\_ABL1| \rmpatient)+(1| \rmrun\_\rmid.\,\rmuniq)\endarray$$
Bulk DNA library preparation, sequencing and read alignment
DNA extracted from peripheral blood was subjected to WGS library preparation and sequenced paired-end, with 150-bp reads on Illumina NovaSeq 6000 machines. The reads were aligned to the human reference genome (GRCh38, NCBI) using the BWA-MEM algorithm.
Bulk unmatched somatic mutation identification and filtering
SNVs were identified using CaVEMan47 for each bulk sample by comparison to an in silico unmatched sample (PD38Is_wgs). CaVEMan was run with the ‘normal contamination of tumour’ parameter set to zero, and the tumour and normal copy numbers set to five and two, respectively. To increase sensitivity, SNVs only flagged as seen in a panel of normal samples (‘VUM’) were rescued. All SNVs were required to have less than half of supporting reads clipped (CLPM = 0) and a median BWA-MEM alignment score greater than or equal to 140 (ASMD ≥ 140). Short insertions and deletions (indels) were called using cgpPindel48 with standard WGS cgpPindel VCF filters applied, except the F010 Pindel filter as it excludes variants seen in a panel of normal samples. Driver candidate variants were restricted to the 37 genes described above. To filter germline variants, we retained SNVs and indels with a gnomAD v.3.1.2 (ref. 56) popmax allele frequency less than 0.01 (annotated using echtvar v.0.2.0, ref. 57). The union of SNVs and indels was taken and reads counted across all samples belonging to the individual using VAFCorrect as above. Sequencing read depth was reviewed in IGV54 to identify arm-level copy number events. GRIDSS v.2.13.1 was used for structural variant calling, and to estimate the VAF of BCR::ABL1.
Bulk phylogeny reconstruction of PD60243
We used DPClust58,59 to infer mutational clusters using SNVs (CaVEMan) and copy number and/or sample purity (Battenberg) called for each sample of PD60243a (81.3 years granulocytes, PD60243c 81.7 years mononuclear cells, PD60243d 81.7 years granulocytes) using a matched buccal sample (PD60243e). Manual inspection of the initial clusters identified 19 outlier poor quality variants that were subsequently removed for a second DPClust run with the addition of all ABL1 mutations identified in the unmatched analysis. All four mutually exclusive ABL1 mutations (as determined by read phasing) were grouped in the same cluster meaning we could not discern the exact subclonal phylogeny of ABL1 mutations (which is expected with bulk reconstruction given the low VAF and 80× coverage). We inferred the total ABL1 mutation burden at blast phase by adding the mutually exclusive ABL1 mutations as individual subclones and removing their assigned cluster. Using ctree (https://github.com/caravagnalab/ctree)60, we performed an exhaustive tree search (sspace.cutoff of 100,000) and retained the top scoring trees that maintained ABL1 mutation mutual exclusivity. We calculated the sum of ABL1 cancer cell fractions (CCF), accounting for nesting, to obtain the CCF median and range.
All of Us cohort analysis
The All of Us Research Program29 is a US population-based cohort that links electronic health record data to WGS data for 206,173 participants, with an average genome-wide coverage of 30×. We searched CRAM files (Data repository v.7) to identify sequencing reads supporting a canonical BCR::ABL1 (or reciprocal ABL1::BCR) variant between BCR (chr. 22: 23289313–23292813) and ABL1 (chr. 9: 130674613–130874613). Reads that were incorrectly oriented or had insufficient mapping quality (score less than or equal to 6) were filtered. GRASS (https://github.com/cancerit/grass) was used to annotate sequencing reads that passed filters. GRASS takes pairs of genomic coordinates representing potential rearrangement events and predicts the fusion consequences along with their associated genes. We required the fusion event to be specifically between BCR and ABL1. After identifying BCR::ABL1 carriers in the cohort, we extracted electronic health record data and searched for International Classification of Diseases (ICD) codes related to cancer, blood or immune diseases. In addition to searching for ‘chronic myeloid leukaemia’ (CML), we included conditions such as ‘abnormal white cell count’, ‘basophilia’, ‘splenomegaly’, ‘unspecified cancer’ or ‘anaemia’. On the basis of these disease entries and domain knowledge, we categorized carriers into four groups: (1) CML mentioned, (2) abnormal white count (for example, leucocytosis), basophilia or splenomegaly mentioned, (3) other haematological issues (for example, anaemia) or unspecified cancer and (4) no relevant disease. Blood sampling date and the date of the most recent ICD code was used to define the time from sample collection to last follow-up. When calculating the incidence of CML and BCR::ABL1 carrier status across age groups, we defined age at diagnosis for CML and age at blood sampling for BCR::ABL1 carrier status. For each age group, summary statistics were calculated by averaging annual statistics from 2018 to 2022, assuming population structure remained stable during this period.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.