Mice
Male and female mice were used for the study. CD45.1+ OT-I or P14 TCR-transgenic mice were housed together. CD45.2+ male and female C57BL/6N or C57BL/6JNifdc mice aged 6–8 weeks were purchased from Vital River as recipients. Female NCG (NOD/ShiLtJGpt-Prkdcem26Cd52Il2rgem26Cd22/Gpt) mice aged 6–8 weeks were purchased from GemPharmatech. Rosa26-Cas9 mice were provided as a gift from the W. Sheng laboratory at the University of Zhejiang. We crossed Rosa26-Cas9 mice with OT-I transgenic mice to generate Cas9+ OT-I mice for CRISPR–Cas9 screening in tumour antigen-specific CD8+ T cells. Four-week-old Klhl6+/− mice were purchased from Cyagen. The Klhl6+/− mice were crossed with OT-I, P14 or C57BL/6N mice to generate Klhl6−/− OT-I/P14 mice or Klhl6−/− mice for subsequent experiments. All mice were kept in a specific-pathogen-free facility, and all animal experiments were performed with the approval of the Institutional Animal Care and Use Committee of Suzhou Institute of Systems Medicine (ISM-IACUC-0151-R and ISM-IACUC-20240098). Mice were housed in standard conditions, with 12 h/12 h light/dark cycles, a controlled temperature of 22–24 °C and humidity of 60%, with unrestricted food and water availability, and were examined daily. All mice were used at 6–16 weeks old. All tumour burdens did not exceed the permission of the Institutional Animal Care and Use Committee of Suzhou Institute of Systems Medicine. Age-matched and sex-matched mice were assigned randomly to experimental and control groups.
Cell lines
Human embryonic kidney 293T (HEK293T) cells were purchased from the American Type Culture Collection (ATCC, CRL-3216) and maintained in DMEM (Gibco, C11995500BT) supplemented with 10% fetal bovine serum (FBS) (Gibco, 16000044) and 1% penicillin–streptomycin (P/S) (Gibco, 15140122). The mouse melanoma cell line B16 was transduced to express OVA257-264 antigen (a gift from Bo Huang laboratory) and maintained in DMEM with 10% FBS and 1% P/S. HepG2 cells (ATCC, HB-8065) were transduced to express human NY-ESO antigen (HepG2-ESO) and cultured in DMEM with 10% FBS and 1% P/S. Jurkat (ATCC, TIB-152) and EL4 (ATCC, TIB-39) cell lines were cultured within the complete Roswell Park Memorial Institute (RPMI)-1640 medium supplemented with 10% FBS, 1% P/S, 1% GlutaMAX (Gibco, 35050061), 10 mM HEPES (Gibco, 15630130), 1% non-essential amino acids (Gibco, 11140076), 1 mM sodium pyruvate (Gibco, 11360070) and 50 μM β-mercaptoethanol (Sigma, M6250). HEK293T, Jurkat, HepG2 and EL4 cells were pre-authenticated by ATCC by short tandem repeat (STR) sequencing. B16-OVA cells were frequently monitored based on their morphological features but have not been authenticated by STR. All cell lines were routinely tested for mycoplasma contamination.
Plasmids
Mouse Klhl6, Tox and Ppargc1a genes were amplified from the complementary DNA (cDNA) library of mice OT-I T cells, and human KLHL6 and TOX genes were amplified from human peripheral blood mononuclear cells (PBMCs). Retroviral plasmid (MSGV-Thy1.1-Klhl6, MSGV-Thy1.1-Ppargc1a and MSGV-Thy1.1-Vector) and packaging vector (pCL-Eco) plasmid were used to produce retroviruses in HEK293T cells using 293 Transfection Reagent (Mirus, MIR 2700), which were then transduced into OT-I CD8+ T cells. The MESV-shCtrl-GFP (Addgene, 85587) was used for Tox or Pgam5 KD. Primer sequences used for Tox and Pgam5 KD can be found in Supplementary Table 6. The retroviral plasmids (MSGV-NGFR-KLHL6, MSGV-Thy1.1-1G4 TCR and MSGV-NGFR-Vector) and packaging vectors (pHIT60 and RD114) were used to produce retroviruses, which were used to transduce PBMCs and Jurkat cells. Lentivirus vectors (pCCLc-MND-Thy1.1-Klhl6 and pCCLc-MND-Thy1.1-Vector) and packaging vector (PA2X and VSV-G) plasmids were used for lentivirus production in HEK293T cells using Liposomal Transfection Reagent for transduction into EL4 cell line. For transient expression experiments in HEK293T cells, the vector plasmid pcDNA4/TO or pFLAG-CMV-4 was used according to the experimental need.
Primary mouse T cell isolation, viral transduction and culture
Naive OT-I T lymphocytes were isolated from the spleens and peripheral lymph nodes of male and female OT-I mice (6–8 weeks). Spleens and peripheral lymph nodes were collected, and mashed through a 70-μm filter, and red blood cells were lysed using red blood cell lysis buffer (BioLegend, 420301) followed by washing with 1× phosphate-buffered saline (PBS). CD8+ OT-I T cells were purified using a CD8+ Naive T cell isolation kit (BioLegend, 480043) according to the manufacturer’s instructions. Primary mouse T cells were counted and then resuspended in RPMI-1640 supplemented with 10% FBS, 1% sodium pyruvate, 1% non-essential amino acids, 10 mM HEPES, 1% GlutaMAX, 1% P/S, 50 μM β-mercaptoethanol and mouse IL-2 (20 U ml−1, Peprotech, 212-12). Then, the resuspended CD8+ OT-I T cells were seeded at a concentration of 1 million cells per ml on 24-well plates with overnight-bound anti-mouse CD3 (2 μg ml−1, BioLegend, 100359) and anti-mouse CD28 (1 μg ml−1, BioLegend, 102121) antibodies. Cells were activated in 24-well plates for 48 h and then transferred out of the activation plates and passaged to new plates every 2 days with a concentration of 1 million cells per ml. For drug treatment experiments, DMSO (Sigma, D2650), 2 μM LFHP-1c (MCE, HY-139598) or 10 μM Mdivi-1 (Selleck, S7162) and 20 μM M1 (Selleck, S3375) were added to cultures daily starting on day 3 after T cell activation. In viral transduction, 7.5 × 105 OT-I cells were transduced with unconcentrated retroviral supernatant after 24 h of activation in 24-well plates coated with RetroNectin reagent (15 μg ml−1, Takara, T100B). Following centrifugation at 2,500 rpm for 90 min at 30 °C, T cells were cultured in the incubator for 24 h. The transduction was repeated 24 h later and then returned to fresh medium for culture. Drug-treated or retrovirus-transduced OT-I cells were sorted by flow cytometry and then adoptively transferred into recipient mice that were inoculated with B16-OVA tumour cells before transfer.
Human T cell isolation, viral transduction and culture
Human PBMCs from healthy donors were purchased from Sailybio and isolated using Lymphoprep (Cytiva, 17144003) according to the manufacturer’s protocol. Isolated PBMCs were cultured in RPMI-1640 medium supplemented with 5% Human Serum AB (Gemini, 100-512), 1% GlutaMAX, 1% non-essential amino acids, 1% P/S, 1 mM sodium pyruvate, 10 mM HEPES and 50 μM β-mercaptoethanol in the presence of human IL-2 (100 U ml−1, Peprotech, 200-02). PBMCs were activated by anti-human CD3 (1 μg ml−1, BioLegend, 317347) and anti-human CD28 (1 μg ml−1, BioLegend, 302943) monoclonal antibodies for 2 days and then underwent viral transduction. In brief, 1 × 106 PBMCs were transferred to a new 24-well plate and dually transduced by 1G4 TCR-specific and KLHL6-specific retroviral supernatant in the presence of 10 μg ml−1 polybrene (Sigma, TR-1003-G). Following centrifugation at 2,500 rpm for 90 min at 30 °C, PBMCs were cultured in the incubator for 24 h with fresh medium and then underwent repeated transduction. The transduced PBMCs were adoptively transferred into female NCG mice that were inoculated with HepG2-ESO tumour cells before transfer.
B16 tumour model and ACT immunotherapy
To investigate the anti-tumour activity of T cells in vivo, 2 × 105 B16-OVA melanoma cells were subcutaneously injected into female C57BL/6N mice. Nine days after tumour implantation, each tumour-bearing mouse was intravenously injected with the required number of CD8+ OT-I T cells from female OT-I mice, which had been expanded for 6 days according to different experimental designs. Tumour-bearing mice received 5 Gy of sublethal irradiation for lymphodepletion 1 day before ACT. For the analysis of tumour growth and mice survival, tumour volume was measured every 2 days and calculated as length (mm) × width (mm) × width (mm) × 0.5. Mice with tumour volumes greater than 1,500 mm3 were euthanized and defined as dead for survival analysis. For the analysis of functional phenotype, mice were euthanized and tissues from tumours, spleens and lymph nodes were collected at days 7, 14, 21 or 28 post-ACT, depending on different experimental designs. For the CellTrace Violet labelling assay, equal numbers of CellTrace Violet-labelled control and KLHL6-OE OT-I T cells were cotransferred into recipient tumour-bearing mice, and TILs were analysed by flow cytometry on day 4 post-ACT. The tumours were digested by Type II collagenase (Worthington Biochemical, LS004176) and processed with Percoll (Cytiva, 17089109). Adoptively transferred OT-I T cells were isolated from tumours, spleens, and lymph nodes, and cell numbers were counted. Isolated T cells were washed and resuspended in ice-cold PBS with 2% FBS in the presence of specific antibodies for the determination of their proportion and functional phenotype through flow cytometry.
In vivo Tpex transfer assay
Female C57BL/6N (CD45.2+) mice were subcutaneously implanted with 2 × 105 B16-OVA cells on day 0. On day 9, each tumour-bearing mouse was intravenously injected with 3 × 106 control or KLHL6-OE CD45.1+ OT-I T cells. Then 14 days after ACT, Tpex (Ly108+TIM-3−) TILs were sorted from tumours by flow cytometry. After sorting, the cells were centrifuged and resuspended in PBS. A total of 5 × 104 Tpex cells were transferred through tail vein injection into female C57BL/6N (CD45.2+) mice that had been subcutaneously implanted with 3 × 105 B16-OVA cells 2 days before. Tumour sizes were measured on day 8 after ACT and every 2 days thereafter. TILs were isolated at days 8 and 16 for phenotypic analysis as previously described in ref. 33.
NCG mouse model and 1G4 TCR-T cell therapy
Female NCG mice were subcutaneously implanted with 4 × 106 HepG2-ESO cells. Subsequently, 1G4 TCR-T cells transduced with or without KLHL6, respectively, were expanded for 12 days in vitro and adoptively transferred into the tumour-bearing mice (6 million cells per mouse) when tumour volumes reached 80 mm3. Mice were euthanized on day 16 after ACT, and the tumours were collected for weighing. For the in vivo phenotyping, the blood, tumours and spleens were collected. The spleens and blood were mashed and/or lysed with red blood cell lysis buffer for 5 min on ice. To isolate T cells from the tumour, the tumours were digested by Type II collagenase and processed with Percoll. Then, the isolated T cells were stained with antibody cocktails and analysed by flow cytometry.
LCMV infection and adoptive T cell transfer
CD45.2+ C57BL/6 recipient mice were intraperitoneally infected with 2 × 105 plaque-forming units (PFU) of LCMV-Armstrong or intravenously injected through the tail vein with 2 × 106 PFU of LCMV-Clone 13. One day before infection, mice received adoptive transfers of 5 × 104 (for Armstrong) or 5 × 103 (for Clone 13) P14 CD8+ T cells. Phenotypic analyses were performed at various time points p.i. according to the experimental design20. Naive WT CD8+ T cells and Klhl6−/− (KO) CD8+ T cells for transfer were isolated from P14 mice using a naive CD8+ T cell isolation kit and adoptively transferred into recipient mice. For retroviral transduction, naive P14 CD8+ T cells were activated for 24 h and then transduced with MSGV-Thy1.1-Klhl6 (KLHL6-OE) or MSGV-Thy1.1-Vector (Control) retrovirus. The following day, transduced CD8+ T cells were sorted, resuspended in cold 1× PBS and adoptively transferred into recipient mice, followed by LCMV infection 1 day later.
LCMV viral RNA quantification
CD45.2+ C57BL/6 recipient mice were intravenously injected with 2 × 106 PFU of LCMV-Clone 13. One day before infection, mice were adoptively transferred with 1 × 104 P14 CD8+ T cells. Liver and lung samples were collected on day 15 p.i., and viral load was quantified using a quantitative PCR (qPCR)-based assay, as previously described60. In brief, total RNA was extracted using the Qiagen RNA isolation kit and subsequently subjected to reverse transcription with the Reverse Transcription Kit (Vazyme, R323-01). cDNA was then used as template for qPCR with 2× SYBR Green qPCR Master Mix (Bimake, b21203). Primers for LCMV GP and hypoxanthine-guanine phosphoribosyltransferase (HPRT) are listed in Supplementary Table 6.
Surprisal analysis
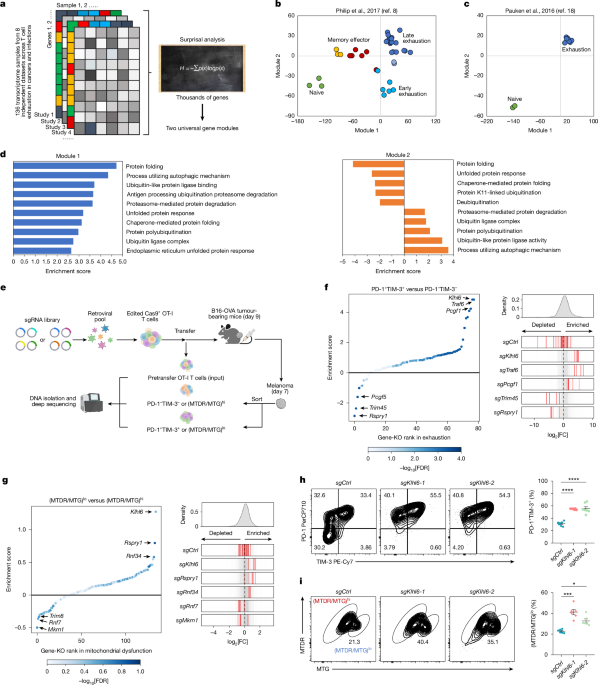
We analysed harmonized bulk RNA-seq datasets comprising 136 samples from 8 previously published studies8,18,33,61,62,63,64,65 including gene expression profiles from CD8+ memory and effector T cells, TILs and chimeric antigen receptor T cells, as well as endogenous Tex cells exposed to chronic antigen stimulation, with or without immune checkpoint inhibition. A complete list of datasets is provided in Supplementary Table 1. Despite thousands of genes that could all be changing across various studies and conditions, we proposed that many genes are coordinately changing together as a group (or gene module), which reflects the fundamental biology of T cell exhaustion programs. Surprisal analysis has been well-documented in deconvoluting the change of thousands of genes into the change of only a couple of gene modules and one unchanged gene expression baseline66,67. The unchanged gene expression baseline reflected the biological processes that are conserved across conditions and time points. The gene module reflected the deviation from the global stable state.
We used surprisal analysis66,67, an information-theoretical analysis technique that integrates principles of thermodynamics and maximal entropy, here to simplify the transcriptome changes into two main gene modules and one unchanged gene expression baseline, which when added together, accurately capture the global transcriptomic profiles of the raw data. Briefly, the logarithm of the measured level of a transcript i at a specific study a sample b, \(\mathrm{ln}{X}_{i}(a\_b)\), is expressed as a sum of a log-transformed gene expression baseline, term \(\mathrm{ln}{X}_{i}^{0}\), and several gene modules \({\lambda }_{j}(a\_b)\times {G}_{{ij}}\), representing deviations from the common expression baseline. Each deviation term is a product of a study-sample-dependent module score\({\lambda }_{j}(a\_b)\), and the study-sample-independent module-specific contribution score Gij of the gene i. Gene i that shows large positive or negative contribution to a module j (high positive or negative Gij value) represents a gene that is functionally positively or negatively correlated with the module j. In other words, the biological function of module j could be inferred by functional enrichment analysis of genes with positive and negative Gij values. The study-sample-dependent module scores of the top modules (in this case, modules 1 and 2) should be able to illustrate the global transcriptome similarities or dissimilarities. To calculate these gene modules, we first computed the singular value decomposition of the matrix \({\rm{ln}}X(a\_b)\). As described previously67, the singular value decomposition factored this matrix in a way that determined the two sets of parameters that are required in the surprisal analysis: the Lagrange multipliers (\({\lambda }_{j}(a\_b)\)) for all gene modules at a given sample and for all samples in all studies, as well as the module-specific contribution scores (Gij for all transcripts i at each gene module j. Further enrichment analysis of the functions associated with each module was performed based on the module-specific contribution scores of the genes associated with that module. These two dominant gene modules (modules 1 and 2) each consist of two gene sets showing opposite expression trends across all samples (M+/−) (Supplementary Table 2). M1+ genes were low in naive and memory T cells but elevated in both early and late exhausted T cells from tumours, as well as in exhausted T cells from chronic infections, whereas M1− genes showed the inverse trend. M2+ genes were selectively enriched in late exhausted cells across the tumour and chronic infection settings, and M2− genes were more highly expressed in naive and early exhausted states.
GSEA of the gene modules
GSEA was performed using MSigDB (v.7.5.1) pathways and custom gene sets derived from the existing literature33,68. Genes were ranked by surprisal analysis scores and analysed separately for association with modules 1 and 2 using the R package clusterProfiler (v.4.12.0)69. Ties (zero scores) were excluded. Enrichment scores were normalized by use of permutation tests, and P values were derived accordingly. Custom gene sets consisted of the top 400 differentially expressed genes (Mann–Whitney U-test) after removing housekeeping, ribosomal and mitochondrial genes. The full GSEA results are provided in Supplementary Table 3.
Mitochondrial function analysis from public databases
To investigate genes associated with mitochondrial function in T cells, we analysed roughly 400,000 TILs from 316 patients across 21 cancer types22, correlating gene expression with pathway activity for hallmark_oxidative_phosphorylation in the Molecular Signature Database. This analysis identified 286 E3 ligases positively associated with mitochondrial function. To account for patient variability in the TIL atlas, we also analysed a ‘cleaner’ mouse RNA-seq dataset within the context of adoptive T cell therapy, in which tumour-specific T cells were sorted into two subsets: (MTDR/MTG)hi functional mitochondria and (MTDR/MTG)lo dysfunctional mitochondria6. Differential pathway enrichment analysis confirmed that ubiquitin-related pathways are significantly associated with mitochondrial function. Through this, we identified 191 E3 ligases positively linked to mitochondrial function. The 133 E3 ligases identified as overlapping between human and mouse analyses (Supplementary Table 4) were selected for in vivo CRISPR screening.
CRISPR–Cas9 screens using the retroviral E3-related library
Retroviral sgRNA vector and sgRNA cloning
In this study, CRISPR–Cas9 sgRNA was expressed using pSL21-Thy1.1 or pSL21-mCherry (Addgene, 164410)23. sgRNAs were generated by annealing two DNA oligos and then ligated into the pSL21-Thy1.1 or pSL21-mCherry vector after digestion with BbsI.
E3-related library construction
The pSL21-mCherry vector was used for the construction of sgRNA library. A computational-guided sgRNA library targeting 78 exhaustion-related E3 ligase genes and 133 mitochondrial-related E3 ligase genes were selected and synthesized. The guide RNA (gRNA) sequences were designed according to previously published data and using the gRNA-design tool (GenScript)70. The library associated with exhaustion differentiation comprised a total of 400 gRNAs, including 10 non-targeting controls and 390 unique sgRNAs, with 5 gRNAs designed for each targeting gene. Another library related to mitochondrial function included a total of 671 gRNAs, including 17 non-targeting controls and 654 unique sgRNAs, with 3–5 gRNAs designed for each targeting gene. All sgRNA oligos, including both positive and negative control sgRNAs, were synthesized by SYNBIO Technologies and pooled in equal molarity. The pooled sgRNA oligos were subsequently amplified through PCR and cloned into BbsI-digested pSL21-mCherry vector using Gibson Assembly Kit (NEB, E5510S). The product of Gibson Assembly reaction was then introduced into TG1 Electrocompetent Cells (Biosearch Technologies, 60502) by means of electroporation and cultured overnight on solid Luria-Bertani agar plates (24 × 24-cm culture plate). The total number of colonies across all plates was counted, exceeding 50× representation, and the plasmids were purified using the EndoFree Plasmid Maxi Kit (CWBIO, CW2104M). To verify the identity and relative representation of sgRNAs in the pooled plasmids, a deep-sequencing analysis was performed by a NovaSeq 6000 (PE150) instrument. We confirmed that 100% of the designed sgRNAs were cloned into the vector and the final library is diverse with a Gini index of 0.05.
In vivo screening
The in vivo screening approach was conducted following established protocols from previous studies23,71. Briefly, a retrovirus pool containing sgRNAs was generated by cotransfecting the specific library plasmids and a packaging vector (pCL-Eco) in HEK293T cells. After 48 h of transfection, the viral supernatant was collected and stored at −80 °C. Naive Cas9+ OT-I T cells were isolated from spleens and activated using anti-CD3 and anti-CD28 antibodies. At 24 h after activation, Cas9+ OT-I T cells were transduced with the retrovirus library, and the transduction process was repeated after 24 h. The transduction efficiency was assessed based on the fluorescence intensity of mCherry, and it reached roughly 40% by the end of transduction. Following viral transduction, the cells were washed and cultured in the medium supplemented with mouse IL-2 for 4 days to allow for expansion and gene editing. mCherry-positive cells were sorted by flow cytometry. Roughly 2 × 105 (400 gRNAs library) or 3.5 × 105 (671 gRNAs library) transduced Cas9+ OT-I T cells were saved as ‘day 0 input’ (around 500× cell coverage per sgRNA). Subsequently, transduced Cas9+ OT-I T cells (3 × 106) were transferred into Cas9+ hosts bearing B16-OVA melanoma tumours. At day 7 after ACT, non-exhausted T cells (PD-1−TIM-3−) and exhausted T cells (PD-1+TIM-3+) or (MTDR/MTG)hi and (MTDR/MTG)lo cells were sorted using flow cytometry and frozen at −80 °C until genomic DNA extraction. A minimum of 2 × 105 or 3.5 × 105 Cas9+ OT-I T cells per sample were collected for further analysis.
sgRNA library sequencing
Genomic DNA was extracted by using the PureLink Genomic DNA Mini Kit (Invitrogen, K182001) according to the manufacturer’s instructions. Two rounds of PCR were performed by using the PrimeSTAR HS DNA Polymerase (Takara, R045B) to amplify the sgRNAs and attach Illumina adaptors and indexes to barcode each sample. The primer sequences used to amplify sgRNAs for the PCR are as follows: next-generation sequencing forward (F), AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGTATTTCGATTTCTTGGCTTTATATATCTTGT; next-generation sequencing reverse (R), CAAGCAGAAGACGGCATACGAGATATTGGCGTGACTGG AGTTCAGACGTGTGCTCTTCCGATCTGACTAGCCTTATTTAAACTTGCTATGC. Different index sequences were added to distinguish between experimental groups. After each PCR reaction, the PCR products were purified using the AMPure XP beads (Beckman A63881). The library sequencing was performed using the Illumina NovaSeq 6000 (PE150) platform (Novogene).
CRISPR screen data processing and analysis
For data analysis, single-end reads were trimmed and quality filtered using the MAGeCK-VISPR package (v.0.5.5)72 and run using Python (v.3.7.4) and matched against sgRNA sequences from the sgRNA library. Read counts for sgRNAs were normalized by control guides when possible, and when not through median counts values. log2 fold changes were calculated with a 1 × 10−2 pseudo-count to account for zero-count genes and avoid infinite values; these fold changes were used as enrichment differences between DP (PD-1+TIM-3+) cell samples and those of DN (PD-1−TIM-3−) cell samples. The same analyses were also performed between (MTDR/MTG)lo versus (MTDR/MTG)hi cells. Gene-targeting sgRNAs consistently showed enrichment or depletion, whereas non-targeting controls were tightly centred around zero, indicating minimal selection bias. Gene retrieval was 100% across all targets in both screens; sgRNA retrieval was 100% in the exhaustion screen and 99.1% in the mitochondrial fitness screen. The log2 fold-change values for each gene and sgRNA from the CRISPR screens are compiled in Supplementary Table 5.
Experimental workflow in RNA-seq
For the transcriptional profiling of Tex cells, we established a B16-OVA melanoma model to analyse tumour antigen-specific CD8+ T cell exhaustion. Briefly, OT-I CD8+ T cells were activated in vitro using anti-CD3 and anti-CD28 antibodies. On day 9 following the implantation of B16-OVA tumours, 3 × 106 OT-I CD8+ T cells were adoptively transferred to each tumour-bearing mouse. On day 14 after ACT, cells were sorted from tumours and spleens using flow cytometry. PD-1 and TIM-3 were used to label different subsets of exhausted TILs: the PD-1−TIM-3− population, PD-1+TIM-3− population and PD-1+TIM-3+ population. For the transcriptional analysis of adoptively transferred WT and Klhl6−/− CD8+ OT-I T cells in the tumours, Klhl6−/− CD8+ OT-I T cells (CD45.1+) and WT CD8+ OT-I T cells (CD45.1/2+) were mixed in a 1:1 ratio and adoptively transferred into the same B16-OVA tumour-bearing mice. After 14 days, the transferred CD45.1+ and CD45.1/2+ CD8+ T cells were sorted from tumours using flow cytometry and prepared for RNA extraction. Total RNA from the isolated transferred OT-I T cells was extracted using the RNeasy Micro Kit (Qiagen, 74004) following the manufacturer’s instructions and stored at −80 °C for RNA-seq. RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent). Subsequently, the libraries were prepared using the TruSeq RNA sample prep kit (Illumina, FC-122-1001). These libraries were then subjected to sequencing on an Illumina NovaSeq 6000 (PE150) platform, generating roughly 40 million paired-end reads (Novogene).
RNA-seq data processing and analysis
The raw read counts were extracted and then normalized by their library size factors and read and gene lengths using edgeR (v.3.36.0)73, which was then used to calculate differential genes. Detailed information on trimming, alignment and quantification can be found as previously reported74 and further details are available at https://github.com/danielgchen/FH_bulk-RNA-seq_pipeline. In brief, data were trimmed using cutadapt (v.2.9)75, quality checked before and after trimming using FastQC (v.0.11.9), and then mapped and quantified using STAR (v.2.7.7a)76. The pathway enrichment analysis of differentially expressed genes was conducted using clusterProfiler (v.4.12.0)77. GSEA was performed with GSEA (v.4.1.0)68; log2 fold changes were calculated with a 1 × 10−2 pseudo-count to account for zero-count genes and avoid infinite values.
Experimental workflow in scRNA-seq
Activated CD8+ OT-I T cells were transduced with either Vector (MSGV-Thy1.1-Vector) or Klhl6 (MSGV-Thy1.1-Klhl6). Then, these transduced cells were adoptively transferred into B16-OVA tumour-bearing mice at a concentration of 3 × 106 cells per mouse. At day 14 after ACT, OT-I T cells were sorted from tumour samples using flow cytometry. Subsequently, these sorted single cells were encapsulated into droplets, loaded into Chromium microfluidic chips with 30 (v.3) chemistry, and barcoded using a 10× Chromium Controller (10X Genomics). The RNAs from these barcoded cells were subsequently reverse-transcribed, and sequencing libraries were prepared using reagents from a Chromium Single Cell 3′ (v3) reagent kit (10X Genomics), according to the manufacturer’s instructions. Library quantification was performed using the Qubit 3.0 Fluorometer (ThermoFisher Scientific), and library quality was assessed using the 2100 Bioanalyzer with the High Sensitivity DNA kit (Agilent). The NovaSeq 6000 platform (Illumina) was used for sequencing the libraries in 50-base pair paired-end mode.
scRNA-seq data processing and analysis
scRNA-seq analysis pipeline closely follows previously reported methods78,79. Briefly, Droplet-based sequencing data were aligned and quantified by use of Cell Ranger Single-Cell Software Suite (v.7.1.0, 10X Genomics) using refdata-gex-mm10-2020-A as a reference. Cells from each sample were first filtered for cells with 500 or more genes and 1,000 or more counts, then filtered based on (1) fewer than 50,000 counts per cell (library size); (2) fewer than 7,000 detected genes per cell and (3) proportion of mitochondrial gene counts (mitochondrial gene unique molecular identifiers (UMIs)/total UMIs) less than 5%. Doublets were identified through clustering; low-quality, low-count cells were also removed. After quality control-based filtering, Scanpy80 was used to normalize cells by means of counts per million normalization (UMI count per cell was set to 106) and log1p transformation (natural log of counts per million plus one). Principal component analysis was performed using variable genes. Leiden clustering and UMAP plots were generated based on selected principal component analysis dimensions. Normalized data are shown in the form of UMAP colour-coding or violin plots. Embedding density was used for density plots and calculated using scanpy.tl.embedding_density, which is a wrapper for the gaussian density algorithm under scipy. TOX signature was defined by taking the genes differentially upregulated, defined as a false discovery rate less than 0.05 and log2 fold change greater than or equal to 1, on Tox-overexpressed T cells in a tumour model38. Stemness and terminal exhaustion signature were defined as the following Lef1, Tcf7, Aqp3, Ccr7, Sell, Il7r, Gzmk, Dusp1, Dusp2, Fos and Junb for stemness and Pdcd1, Ctla4, Cd200r1, Cd244a, Havcr2, Lag3 and Tigit for terminal exhaustion; genes were derived from literature. Published datasets on T cell exhaustion were obtained from studies related to chronic infection and human tumour-infiltrating CD8+ T cells22,33. The public T cell exhaustion data in chronic infection was processed from raw by filtering for n counts between 2,000 and 10,000, n genes between 1,000 and 3,000, and less than 5% mitochondrial reads; data were then normalized according to the aforementioned methods and projection algorithms.
qPCR with reverse transcription
Total RNAs from cells were extracted using Trizol reagent (Takara, 9109) or the RNeasy Micro Kit, according to the manufacturer’s instructions. The extracted RNAs were reverse-transcribed into cDNA using HiScript Reverse Transcriptase (Vazyme, R323-01). Quantitative real-time PCR was performed using the ABI prism 7500 real-time PCR System (ThermoFisher) and 2× SYBR Green qPCR Master Mix (Bimake, b21203), following the respective manufacturer’s protocols. The data are presented as the fold change in gene expression normalized to an internal reference gene (B2M). The relative expression of mRNA was calculated using the 2−ΔΔCT method. Primer sequences used for qPCR can be found in Supplementary Table 6.
Flow cytometry and sorting
T cells were stained using fluorescent antibodies and subsequently analysed by flow cytometry. To prepare for staining, T cells were collected and washed once with fluorescence-activated cell sorting (FACS) buffer (PBS with 2% FBS). For surface protein staining, cells were stained with fluorescently conjugated antibodies and Live/Dead Fixable Dead Cell Stain Kit (Invitrogen, 65-0866-18) in FACS buffer, then fixed with 2% paraformaldehyde (Casmart) for 30 min at 4 °C. For transcription factor staining, cells were prestained with Live/Dead Fixable Dead Cell Stain Kit and fluorescently conjugated antibodies in FACS buffer to detect surface markers. The cells were then fixed for 30 min at 4 °C using FOXP3/transcription factor fixation buffer (Invitrogen, 00-5523-00) and stained with transcription factor antibodies in permeabilization buffer (Invitrogen, 00-8333-56). For detection of intracellular cytokines, cells were stimulated with phorbol myristate acetate in the presence of Brefeldin A (BFA) (BioLegend, 423304) for 4.5 h. Then, the prestained cells were fixed and stained with cytokines antibodies in the permeabilization buffer. After staining, cells were resuspended in FACS buffer for flow cytometric analysis. Flow cytometry data were collected using BD LSR Fortessa and BD FACSDiva (v.8.0.2), and analysed with FlowJo (v.10.4) software. Cell sorting was performed using BD FACS Aria III and BD FACSDiva (v.8.0.2). A list of antibodies and their dilutions used can be found in Supplementary Table 8.
Transmission electron microscopy
The indicated OT-I T cells were activated in each well of a 24-well plate with mouse anti-CD3 and anti-CD28 antibodies for 3 days and cultured in RPMI-1640 medium for another 3 days. Subsequently, 1 × 106 OT-I T cells were gathered and fixed in a precooled fixation buffer (2.5% glutaraldehyde, 0.1 M phosphate buffer, pH 7.4) overnight at 4 °C. After 3 washes with PBS, cells were postfixed in 1% osmium tetroxide in PBS for 2 h, dehydrated and embedded in Spurr’s resin following standard procedures. Ultrathin sections were stained with uranyl acetate and lead citrate. Mitochondrial morphology was visualized using Hitachi HT-7800 transmission electron microscopy (v.01.20) and an AMT-XR81DIR camera. For quantitation of mitochondrial cross-sectional area and total crista length, images of each cell profile containing four or five mitochondria from a single thin section for the indicated samples were analysed. Cross-sectional area and total crista length per mitochondrion were calculated using the lasso tool in Image J (v.1.8.0) software.
Seahorse analysis
To investigate metabolic characteristics, we used a Seahorse XF24 analyser (Agilent) to measure both OCR and glycoPER in in vitro-expanded T cells and TILs sorted from tumours at day 14 after ACT, according to different experimental designs. Before analysis, these cells were pretreated with the non-buffered XF medium (RPMI-1640 supplemented with 10 mM glucose, 1 mM sodium pyruvate and 2 mM glutamine). Subsequently, the cells were seeded at a density of 1.3 × 105 cells per well in an XF24 cell culture microplate and incubated in a non-CO2 environment for 1 h at 37 °C. To optimize cell adhesion, the plates underwent a 5 min spin at room temperature at 100g with zero brake. Measurements of OCR and glycoPER were conducted under both basal conditions and in response to specific compounds, such as 1.25 μM oligomycin (Oligo), 50 mM 2-deoxy-d-glucose, 1.5 μM carbonyl cyanide-p-trifluoromethoxy-phenylhydrazone, 0.5 μM rotenone and antimycin A (R&A). The SRC was calculated by subtracting basal OCR from maximum OCR. The OCR coupled with mitochondrial ATP production (coupled OCR) is defined as the OCR reduction after the injection of oligomycin, which inhibits ATP synthase. OCR and glycoPER were analysed by Seahorse wave software (Seahorse, Agilent Technologies, v.2.6).
Mitochondrial mass and membrane potential analysis
Mitochondrial mass and membrane potential were assessed using MTG (Invitrogen, M7514) and either TMRE (Invitrogen, T669) or MTDR (Invitrogen, M46753). In brief, cells were stained with 250 nM MTG and 50 nM TMRE or MTDR, and incubated at 37 °C (5% CO2) for 30 min. Following incubation, cells were washed three times with FACS buffer and subsequently subjected to surface marker staining for further flow cytometric analysis.
Western blotting
For protein expression analysis, cells were gathered, washed with cold PBS, and then lysed in 1% SDS (Sangon, 151-21-3) for 30 min on ice. The protein samples were denatured at 95 °C for 15 min and stored at −20 °C. Protein samples were separated on SDS–PAGE gels and then transferred onto methanol-activated polyvinylidene fluoride membranes (Millipore, IPVH00005). Membranes were blocked with 5% nonfat milk in PBS containing Tween-20 (0.1%) for 1 h and then incubated overnight at 4 °C with the respective primary antibodies. The following day, membranes were incubated with the corresponding HRP-coupled secondary antibodies for 2 h at room temperature, followed by signal development using ECL Western Blotting substrate (Tanon, 180-5001) and the Chemidoc automated detection system (Bio-Rad). Data analysis was performed using Image J (v.1.8.0) software. The antibodies and their dilutions used can be found in Supplementary Table 8.
E-STUB for mass spectrometry
As previously described in ref. 37, Jurkat cells were cultured in DMEM supplemented with 10% dialysed FBS. Three biological replicates were prepared for each treatment condition. Jurkat cells were transduced with retroviruses packaged in HEK293T cells using MSGV-NGFR-KLHL6-BirA and MSGV-Thy1.1-BAP-Ub plasmids to co-express KLHL6-BirA and BAP-tagged ubiquitin. Cells transduced with retroviruses packaged from MSGV-NGFR-Empty-BirA and MSGV-Thy1.1-BAP-Ub plasmids served as controls. Then 72 h post-transduction, cells were pretreated with proteasome inhibitor MG132 to promote accumulation of ubiquitylated substrates and then pulsed with 50 μM biotin for 30 min. Following biotin labelling, cells were washed once with ice-cold PBS and lysed on ice using E-STUB RIPA buffer (RIPA buffer supplemented with EDTA-free protease inhibitor cocktail, Pierce Universal Nuclease, 10 mM N-ethylmaleimide, 1 mM EGTA and 1.5 mM MgCl2). Lysates were collected into microcentrifuge tubes, rotated at 4 °C for at least 1 h and clarified by centrifugation at 12,000g for 15 min at 4 °C. Total protein lysates were incubated with 50 μl of resuspended and prewashed streptavidin beads overnight at 4 °C with rotation. The following day, beads were washed 5 times with Wash Buffer (PBS containing 0.05% Tween-20), then incubated with 30 μl of 0.1% SDS at 95 °C for 5 min in a metal bath. Samples were sent to Shanghai Omicsspace Biotech for mass spectrometry analysis. Significant changes between the relative protein abundance of the experimental samples to the control samples were assessed by two-sided moderated t-test as implemented in the limma package (v.3.54.2)81 (Supplementary Table 7).
Co-IP and ubiquitination assays
For the Co-IP assay, cells were transfected with the plasmids according to the experimental designs outlined in the figures. After 36–48 h, the cells were gathered and lysed in Co-IP lysis buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 2.5 mM sodium pyrophosphate and 1 mM Na3VO4, for 1 h at 4 °C. The cell lysates were centrifuged at 12,000g for 10 min at 4 °C to remove cell debris. The supernatants were collected and incubated with anti-Flag or anti-Myc beads that had been precleaned with Co-IP buffer. These mixtures were then rotated overnight at 4 °C. On the following day, the beads were subjected to five washes with Co-IP lysis buffer and then resuspended in 1× loading buffer. Subsequently, the samples were denatured at 95 °C for 15 min and stored at −20 °C. To assess the ubiquitination of TOX and PGAM5, TOX proteins (both endogenous and exogenous TOX proteins) and endogenous PGAM5 proteins were immunoprecipitated from cell lysates using anti-Flag beads or TOX/PGAM5 antibody-coated beads, depending on the different experimental conditions. In brief, cells were collected and lysed using ultrasonic cracking, and then denatured at 95 °C for 15 min. Cell lysates were incubated in Co-IP lysis buffer with protease inhibitors (Roche) and rotated for 1 h at 4 °C. Subsequently, the cell lysates were centrifuged at 12,000g for 10 min at 4 °C to collect cell supernatant. The supernatant was incubated with the respective antibody–beads complex and rotated overnight at 4 °C. Afterwards, the beads were washed 5 times with Co-IP buffer and denatured by heating at 95 °C for 15 min. These samples were separated by SDS–PAGE, transferred to polyvinylidene fluoride membranes and then subjected to western blotting using the designated primary and secondary antibodies.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (v.8.0). No statistical methods were used to predetermine sample size. Data collection and analysis were conducted without blinding to the experimental conditions. A two-tailed Student’s t-test was used to compare the two groups. For multiple comparisons, a two-way analysis of variance (ANOVA) with Tukey’s or Sidak’s multiple-comparisons test was applied. The log-rank (Mantel–Cox) test was performed to compare mouse survival curves. Data are presented as mean ± standard error of the mean (s.e.m.). The numbers of mice used per experiment and the number of experimental repeats are indicated in the figure legends. P < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.