Patient samples
All retrospective medical data/biospecimen studies at the Netherlands Cancer Institute have been executed pursuant to Dutch legislation and international standards. Before 25âMay 2018, national legislation on data protection applied, as well as the International Guideline on Good Clinical Practice. From 25âMay 2018 we also adhered to the General Data Protection Regulation of the European Union. Within this framework, patients are informed and have always had the opportunity to object or actively consent to the (continued) use of their personal data and biospecimens in research. Hence, the procedures comply both with (inter)national legislative and ethical standards. All retrospective medical data/biospecimens provided by LUMC were part of the DIRECT study (NCT02126449 (ref. 45)), were conducted in accordance with the Declaration of Helsinki (October 2013) and were approved by the Ethics Committee of LUMC in agreement with Dutch law for medical research involving human subjects.
Assessment of HER2 status in this study followed established guidelines, with scoring based on both immunohistochemistry (IHC) and in situ hybridization techniques. Immunohistochemistry results were categorized as 0/1+ (HER2-negative), 2+ (equivocal; considered positive if amplification was detected with in situ hybridization) or 3+ (HER2-positive). Assessment of oestrogen receptor was performed according to Dutch guidelines, with scores of 10% or above considered positive, and those below 10% negative. Pathological complete response (pCR) scoring adhered to local standard guidelines. For one patient, pCR was determined based on radiological images because surgery was not performed due to pCR having been achieved. For measurement of tumour reduction from baseline, we compared pretreatment radiological images with post-treatment residual disease as assessed by pathological evaluation.
Experimental model and subject data
All mice were adult females, housed under a 12/12âh light/dark cycle and under specific-pathogen-free laboratory conditions and received food and water ad libitum. All experiments were approved and performed according to the guidelines of the Animal Welfare Committees of the Royal Dutch Academy for Sciences (Hubrecht Institute), the Netherlands Cancer Institute or KU Leuven. Sample size was not determined a priori. Mice were randomly assigned to experimental groups. All experiments were performed in a blinded manner, except for tumour volume measurement, which was performed by the same investigator who administered chemotherapy treatment, making it impossible to work in a blinded manner.
MMTV-PyMT26 and MMTV-Cre61 mice were purchased from Jackson Laboratory. E-cad-mCFP mice62 were a gift from H. Clevers, R26-loxP-stop-loxP-YFP (R26R-YFP) mice a gift from J. Deschamps and MMTV-Wnt1 (ref. 30) mice a gift from J. Hilkens. MMTV-PyMT;R26R-Confetti;R26-CreERT2 mice62,63, of a mixed BL6/FVB genetic background, were used to label and trace single cells by IVM. Following tumour development, mice were intraperitoneally injected with tamoxifen (Sigma-Aldrich; 1.5âmgâ25âgâ1, diluted in sunflower oil) to activate Cre recombinase and induce colour randomization of the confetti cassette. MMTV–Wnt1;R26R-Confetti:R26-CreERT2 mice62,63, of a mixed BL6/FVB genetic background, were used to isolate tumour pieces, which were transplanted into NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) female mice (8â12âweeks of age) Following tumour development, mice were intraperitoneally injected with tamoxifen (Sigma-Aldrich; 1.5âmgâ25âgâ1, diluted in sunflower oil) to activate Cre recombinase and induce colour randomization of the confetti cassette. Staining and treatment experiments in transgenic mice were performed on MMTV-PyMT;MMTV-Cre;R26-LSL-YFP;E-cad-mCFP and MMTV-Wnt1;MMTV-Cre; R26-LSL-YFP;E-cad-mCFP mice of a pure FVB genetic background.
For staining and treatment experiments in transplanted models, tumours from three independent, treatment-naive MMTV-PyMT;MMTV-Cre;R26-LSL-YFP;E-cad-mCFP donors were harvested. Mammary tumours were minced and enzymatically digested by gentle shaking for 30âmin at 37âC in digestion mix (0.2% trypsin from bovine pancreas; Sigma) and 0.2% collagenaseâA (Roche)). The digested tumours were spun down and cell fragments embedded in basal membrane extract (BME) (RGF BME typeâ2 PathClear). Mammary tumour organoid medium contained DMEM/F12 GlutaMAX (GIBCO), 2% B27 (Invitrogen) and 10ângâmlâ1 fibroblast growth factor. Brca1â/âTrp53â/â organoids were a gift from J. Jonkers. Both MMTV-PyMT and Brca1â/âTrp53â/â organoids were orthotopically transplanted into both fourth mammary glands of 8â12-week-old FVB/NRj female mice (Janvier Labs).
For transplantation of MMTV-PyMT organoids, 250,000âsingle cells were plated 3âdays before transplantation. On the day of transplantation, BME was dissolved by mechanical disruption and cells dissolved in 100âμl of BME typeâ2 (RGF BME typeâ2 PathClear)/PBS. For transplantation of Brca1â/âTrp53â/â organoids, 10.000âsingle cells were transplanted in both fourth mammary glands of FVB/NRj mice.
Staging of mice
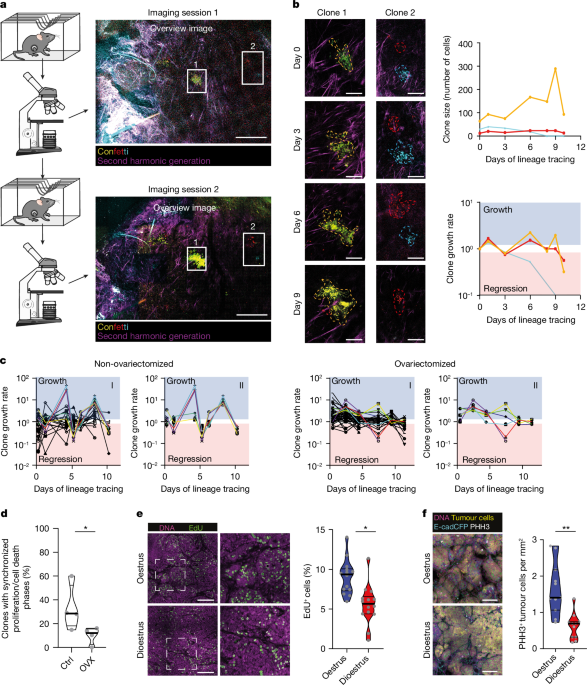
For determination of the oestrous stage of mice, a vaginal smear was collected and analysed as described in refs. 24,46,64. In short, the vagina was flushed with 50âµl of PBS which was then transferred to a glass slide. Following air drying, the slide was stained with 0.01% crystal violet and oestrus stage determined by examination of cytological characteristics using a light microscope. Mice were categorized as regularly cycling when they showed cycles of length 4â6âdays37.
Ovariectomy
In a subgroup of mice, ovariectomy was performed when mammary tumours of volume 50âmm3 could be palpated, after which mice were allowed to recover for at least 7âdays before IVM or treatment.
Chemotherapeutic treatment of mice
When tumours had reached a cumulative volume of 500â750âmm3 following short-term treatment, or either 250â450âmm3 (transgenic MMTV–PyMT model) or 50â200âmm3 (transplanted models) for long-term treatment, mice were treated with either vehicle (PBS) or chemotherapeuticâeither doxorubicin hydrochloride (5âmgâkgâ1; catalogue no. D1515, Sigma-Aldrich) or cyclophosphamide monohydrate (100âmgâkgâ1; catalogue no. C7397, Sigma-Aldrich)âonce per week by intravenous injection. Treatment cycles were continued for a maximum of 5âweeks or until the humane endpoint was reached (cumulative tumour load over 750âmm3 or single tumour over 350âmm3). Tumour diameter was measured three times per week by calipers. Following the last treatment cycle, primary tumours and lungs were collected from these mice for further analyses.
Depletion experiments
When mice approached a cumulative tumour volume of 500â750âmm3, they began receiving depletion antibodies at least 1âweek before chemotherapeutic treatment and this continued until the end of the experiment. Individual mice were treated every week with either 60âmgâkgâ1 of either InVivoMAb anti-mouse CSF1R (CD115) monoclonal antibody (BioXCell, clone AFS98, catalogue no. BE0213) or InVivoMAb rat IgG2a isotype control (BioXCell, clone 2A3, catalogue no. BE0089), or received InVivoMAb anti-mouse CD4 (BioXCell, clone GK1.5, catalogue no. BE003-1) and InVivoMAb anti-mouse CD8 (BioXCell, clone YTSâ169.4, catalogue no. BP0117), three times per week, every other day, with a first dose of 400âµg followed by 200âµg per antibody.
Mammary imaging window implantation and IVM
To follow in vivo tumour clone dynamics, a mammary imaging window was inserted into tumour-bearing mice 2â3âdays following induction of lineage tracing. Mice were anaesthetized using a mixture of 2% isoflurane and compressed air, with surgery performed under aseptic conditions. Before surgery, the skin overlying the tumour was shaved and the skin disinfected using 70% ethanol. An incision was made through the skin overlying the tumour and an imaging window inserted65,66. The imaging window was secured using a non-absorbable, non-woven, purseâstring suture (4â0 prolene suture) in the skin. For imaging, mice were sedated using isoflurane inhalation anaesthesia (roughly 2.0% isoflurane/compressed air mixture) and received 200âµl of sterile PBS by subcutaneous injection to prevent dehydration. Mice were placed in a custom-designed imaging box on the microscope stage and maintained under constant anaesthesia under the microscope within a temperature-adjusted climate chamber (34.5â°C). Following each imaging session, mice were allowed to recover on a heat mat. Imaging was performed on either an inverted Leica TCS SP5 AOBS multiphoton microscope with a chameleon Ti:Sapphire pumped Optical Parametric Oscillator (Coherent Inc.; www.coherent.com) or a Leica SP8-DIVE confocal microscope equipped with an Insight X3 dual-beam pulsed layer (Spectra Physics; www.spectra-physics.com), equipped with non-descanned detectors. CFP was excited at a wavelength of 840ânm, and yellow fluorescent protein (YFP), green flourescent protein and red flourescent protein at a wavelength of 960ânm. All images were in 12-bit and acquired with a Ã25â(HCX IRAPO, numerical aperture 0.95, working distance 2.5âmm) water objective with a free working distance of 2.40âmm. Three-dimensional tile scans of large tumour areas were taken at the indicated time points (at least 5.0âÃâ5.0âÃâ0.5âmm3 (x, y, z)) with z-steps of 5â10âµm. Tile scans were collected at regular intervals over time periods of up to 5âweeks following induction of lineage tracing. The same imaging fields were followed during subsequent imaging sessions, using the imaging coordinates of the first imaging session.
Postprocessing and analysis of IVM data
For analysis of three-dimensional tile scans, all intravital images were stitched and processed using Leica Application SuiteâX software (Leica Microsystems) and ImageJ software (https://imagej.net). Clones were manually annotated in the images, and clone size (number of cells) counted in three dimensions throughout the z-stacks. By following the same clones in our intravital images, we quantified individual clone sizes over time. The fraction of clones that behaved in a synchronized manner with phases of growth, when clones become larger, followed by a phase of regression during which the clones become smaller, was denoted as alternating. To distinguish synchronized clones from non-synchronized in quantitative terms, we identified as synchronized clones those that showed clear, regular alternating phases of growth and regression with the same periodicity, similar to a sinusoidal wave, over multiple consecutive cycles throughout the imaging sessions. By contrast, non-synchronized clones did not exhibit this regular pattern.
Histochemistry and immunohistopathology
Mouse tissues were formalin fixed and paraffin embedded. Haematoxylin and eosin staining was performed on 2âµm sections and IHC staining on 4âµm sections using routine procedures. For IHC staining, antigen retrieval was performed with Tris/EDTA (pHâ9.0) (Tris: Sigma, catalogue no. 252859; EDTA: Sigma, catalogue no. EDS) for ER-α (Invitrogen, clone 6F11, catalogue no. MA1-27107, 1:100), CD4 (Invitrogen, clone 4SM95, catalogue no. 14-9766-82, 1:1,000), CD8 (Invitrogen, clone 4SM15, catalogue no. 14-0808-82, 1:2,000), CD31 (Cell Signaling, catalogue no. 77699S, 1:100), F4/80 (Cell Signaling, catalogue no. 70076S, 1:1,000) or citrate buffer (pHâ6.0) for progesterone receptor (Fisher Scientific, clone SP2, catalogue no. RM-9102-S1, 1:300). Sections were incubated with primary antibodies overnight at 4â°C. For CD4 and CD8 staining, additional labelling was performed by incubation of goat anti-rat biotin (SouthernBiotech, catalogue no. 3052-08, 1:150) antibody for 30âmin at room temperature. Following washing, binding of primary antibody was visualized dependent on species using either the EnVision+ HRP Labelled Polymer anti-rabbit/mouse system (Dako, catalogue nos. K400, K4003 and K4001) or Streptavidin/HRP (Dako, catalogue no. P0397) and the Liquid DAB+ Substrate Chromogen System (Dako, catalogue no. K3468); counterstaining with haaematoxylin was then performed. Slides were digitally processed using a PANNORAMIC 1000 whole-slide scanner (3DHISTECH) and images captured with SlideViewerâ2.7 (3DHISTECH). For determination of positive cells, IHC staining was quantified using QuPathâ0.4.4 (GitHub) with atomized classifiers.
Immunostaining
Tumour samples were fixed in periodate/lysine/4% paraformaldehyde buffer overnight at 4â°C, incubated in 30% sucrose overnight at 4â°C and embedded in Tissue Freezing medium (Leica Biosystems). Tumours were cryosectioned and immunostaining performed on 10âµm sections. For this, sections were hydrated in PBS for 10âmin at room temperature and subsequently blocked and permeabilized for 1âh with 0.5% TritonX/5% normal goat serum (NGS) in PBS. The primary antibody used was anti-PHH3 (Millipore, catalogue no. 06-570, 1:500); the antibody was diluted in 0.1% TritonX/5% NGS in PBS, and tissues were sectioned and stained overnight at 4â°C. Following three washes in PBS, the secondary antibody (donkey anti-rabbit AF568; Invitrogen, catalogue no. A10042, 1:1,000) was incubated for 1âh at room temperature and nuclei stained with TO-PRO-3 (Invitrogen, catalogue no. T3605, 1:5,000). Following three 10âmin washes in PBS, stained sections were mounted using VectaShield. All stainings were imaged with an inverted Leica TCS SP8 confocal microscope. Fluorophores were excited as follows: AF594 at 561ânm, YFP at 514ânm and TO-PRO-3 at 633ânm. YFP was collected at 519â555ânm, Alexa-568 at 575â630ânm and TO-PRO-3 at 644â694ânm. All images were collected in 12-bit with a Ã25âwater-immersion objective (HC FLUOTARâL, numerical apertureâ0.95, working distance 2.4âmm). For determination of positive cells, staining was quantified using Fiji/ImageJ v.1.49k and Excelâ2016.
EdU detection
EdU was injected intraperitoneally 4âh before mice were killed (1âmg per animal, 5âmgâmlâ1 stock in PBS; Sigma, catalogue no. 900584). EdU staining was performed on paraffin sections of thickness 10âµm; sections were deparaffinized, and hot target retrieval performed in citrate buffer pHâ6.0. EdU was detected by incubation with 100âmM Tris pHâ8.5, 1âmM CuSO4, 100âmM ascorbic acid and 10âμM AlexaFluor-488 azide (Invitrogen, catalogue no. A10266) for 30âmin at room temperature. Subsequently, DNA counterstaining was performed with TO-PRO-3 (Invitrogen, catalogue no. T3605, 1:5,000) in 0.1% TritonX/5% NGS in PBS for 1âh at room temperature. Following three 10âmin washes in PBS, stained sections were mounted using VectaShield. All staining was imaged with an inverted Leica TCS SP8 confocal microscopes. Fluorophores were excited as follows: AF488 at 488ânm and TO-PRO-3 at 633ânm; AF488 was collected at 492â530ânm and TO-PRO-3 at 650â700ânm. All images were collected in 12-bit with a Ã25âwater-immersion objective (HC FLUOTARâL numerical aperture 0.95âW, VISIRâ0.17, FWD 2.4âmm). For determination of positive cells, staining was quantified using semiautomated macros in Fiji/ImageJ v.1.49k and Excelâ2016.
Flow-cytometric analysis of dying cells
Mammary tumours were collected separately and minced on ice using sterile scalpels, followed by digestion for depletion experiments in 25âμgâmlâ1 DNaseâI (Roche) and 5âWünsch units TH Liberaseâmlâ1 (Roche) in PBS at 37âC for 35âmin, or in 25âμgâmlâ1 DNaseâI (Roche) and 3âmgâmlâ1 collagenaseâA (Sigma) in PBS at 37â°C for 1âh. The digestion mix was filtered through a 70âµm filter (BD Falcon) while adding DMEM/F12â+âGlutaMAX, followed by spinning down for 4âmin at 500ârelative centrifugal force at 4â°C and resuspension of pellets in 5âmM EDTA/PBS. Cells were washed once in 5âmM EDTA/PBS and centrifuged (4âmin at 500ârelative centrifugal force at room temperature) before proceeding with antibody labelling. Tumour cells were blocked in fluorescent activated cell-sorting buffer supplied with 20% normal goat serum (Gibco) for 10âmin on ice, before labelling with one of the following antibody combinations for depletion experiments: (1) E-cad-eFluor660 (catalogue no. DECMA-1, eBioscience, 1:200), biotin-conjugated anti-mouse CD41 clone eBioMWReg30 (eBioscience, catalogue no. 13-0411-821, 1:200) and anti-mouse CD45 clone 30-F11 (eBioscience, catalogue no. 13-0451-85, 1:200); (2) E-cad-eFluor660 (catalogue no. DECMA-1, eBioscience, 1:200); and (3) anti-mouse CD45-BUV395 clone 30-F11 (BD Bioscience, catalogue no. 564279, 1:200). Secondary labelling was performed using streptavidin-conjugated PerCP (BioLegend, catalogue no. 405213, 1:200). Dead cells were stained using the Apoptosis detection kit PE (Invitrogen, catalogue no. 88-8102-74) according to the manufacturerâs protocol. For depletion experiments no secondary labelling was performed and, following washing, cells were immediately stained with PI and analysed on a SymphonyâA5 (BD Biosciences) then washed once in 5ânM EDTA/PBS and analysed on a Symphony A5 (BD Biosciences). A broad forward scatter/side scatter (FSC/SSC) gate was followed by gates excluding doublets. Immune cells and megakaryocytes were then excluded, based on staining for CD41 and CD45, in a dump channel. Tumour cells were selected according to YFP positivity and further stringently gated for the presence of the cell death marker annexin and PI, or for PI only for depletion experiments. Data were manually analysed with FlowJo v.10.6.2. Analysis of staining and fluorescent activated cell-sorted samples was pseudomized by number coding.
RNA isolation, complementary DNA preparation and qPCR
RNA was isolated using Trizol reagent (Invitrogen Life Technologies) according to the manufacturerâs protocol. Purity and amount of RNA isolated were analysed using a Nanodrop spectrophotometer. Complementary DNA was prepared using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturerâs protocol. Sequences of used primers can be found below. Quantitative PCR (qPCR) was performed using Power SYBR Green PCR Master Mix (Applied Biosystems). Thermal cycle conditions used for all qPCR reactions were as follows: 5âmin at 95â°C, followed by 40âcycles of denaturation for 30âs at 95â°C, annealing for 30âs at 60â°C and extension for 1âmin at 72â°C. PCR reactions were concluded with incubation for 10âmin at 72â°C to complete the extension of all synthesized products.
Progesterone assay
To 250âµl serum or calibrator samples was added 10âµl of deuterated internal standard (progesterone-d9, CDN Isotopes). Subsequently, progesterone was extracted with 1âml of methyl tert-butyl ether and dried using a SpeedVac concentrator. Dried samples were reconstituted in 100âµl of injection solution (methanol:water 2:3, v:v). Thereafter, samples were centrifuged for 5.0âmin at 18,213g and 50âµl was injected into a Nexera SIL30ACMP (Shimadzu) autosampler. Chromatographic separation was achieved using a Kinetex EVO 1.7âµm C18 column (2.1âmm, internal diameter 50âmm) (Phenomenex). A gradient protocol of two mobile phases, containing water with 0.1% formic acid and 2âmM ammonium acetate, and methanol, was applied at a flow rate of 0.6âmlâminâ1. The gradient started at 50% methanol, which was gradually increased to 75% over 2.3âmin. Next, the column was washed with 100% methanol for 0.5âmin before returning to the starting condition of 50% methanol, for 0.7âmin. The assay had a total run time of 3.5âmin. Tandem mass spectrometry analysis was performed with a QTRAP6500+ instrument (Sciex) operated in positive electrospray ionization mode (600â°C) and multiple-reaction monitoring mode. For quantitation of progesterone, mass:charge ratio, 315.1âââ97.0 (progesterone) and 324.2âââ100.0 (progesterone-d9) were monitored. The assay is standardized against the NIST SRMâ971 standard. In two serum pools containing 1.1 and 27.5ânmolâlâ1, total coefficient of variation was 9.5 and 7.5%, respectively. The lower limit of quantitation was determined at 0.96ânmolâlâ1 (coefficient of variation 8.3%) in a serum pool containing low progesterone levels. Analysis of patient samples (sera) was performed blinded, and linked to tumour volume measurements and other clinical parameters only at a later stage.
Statistics and reproducibility
Statistical analyses were performed using Râv.4.4.2 (R Development Core Team and the R Foundation for Statistical Computing) by integration of software from open-source packages. For further details on statistics see Supplementary File 1. The level of statistical significance was set at #Pâ<â0.1, *Pâ<â0.05, **Pâ<â0.01, ***Pâ<â0.001. For all violin plots, thicker solid centre lines represent the median, thinner solid lines the 25th and 75th percentiles. Statistical analyses of longitudinal mouse data were performed using Râv.4.4.2 (R Development Core Team and the R Foundation for Statistical Computing) by integrating software from open-source packages, including nlme67, JMbayes2 (ref. 68) and packages from tidyverse69, including dplyr, tidyr and ggplot2, and were analysed as follows. Both tumour volume over time and time to event (either end of study or death) were recorded for each individual animal. To test for survival, survival regression (Cox) models were used to test whether survival probability between groups was different. For longitudinal tumour measurements, the mixed-effects model70 was used. Regression splines (natural cubic splines with 2 or 3 degrees of freedom, depending on the case) were used to fit a nonlinear tumour growth function per animal along time, taking into account all separate tumours for each individual animal separately. This model takes the correlated structure of the data into consideration by inclusion of individual animals as a grouping factor, and tumour (within the animal) as a subgrouping factor. Furthermore, the effects of time and treatment were considered as random and fixed effects, respectively. A joint model was used to combine survival and tumour volume measurements, to determine whether the responses together were different between treatment groups. Splines were again used to represent the effect of time. For this model, multiple tumours per animal had their growth curves averaged. For further details on these statistics, see Supplementary File 2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.