Cell lines
The KP cells used here were established previously66. Atf4 and Lcn2 knockout cell lines were generated by transient transfection of PX458 (Addgene, 48138) expressing guides targeting either Atf4 or Lcn2. The list of sequences and primers is included in Supplementary Table 4. Single GFP-positive clones were selected, and gene of interest (GOI) loss was validated by western blot or ELISA. B16F10 cells were a gift from the I. Aifantis laboratory. Cells were cultured in RPMI with 10% fetal bovine serum (FBS) and gentamicin. mLCN2 and hLCN2 KP cells were selected and maintained in hygromycin-supplemented medium (800 μg ml−1). Commonly available cell lines have been authenticated using short tandem repeat (STR) profiling. All cells were mycoplasma-negative.
The mouse KPC7 pancreatic cancer cell line, derived from a tumour in LSL–KrasG12D/+;LSL–Trp53R172H/+;Pdx1-Cre mice, was used. This cell line is maintained on a C57BL/6 background.
Doxycycline-induced knockdown of Lcn2 was achieved by cloning hairpin targeting Lcn2 into the pLKO.1-TETON-Puro vector (Addgene plasmid 21915). In brief, hairpins were designed according to the Genetic Perturbation Platform (Broad Institute), and the annealed hairpin is ligated into the EcoRI+AgeI–digested backbone with Quick Ligase (NEB) at a 3:1 insert:vector molar ratio. Sequences and primers are listed in Supplementary Table 4. Vectors were transduced into cells through lentivirus and selected with puromycin (8 μg ml−1). Oligos were obtained from Integrated DNA Technologies.
Mouse models
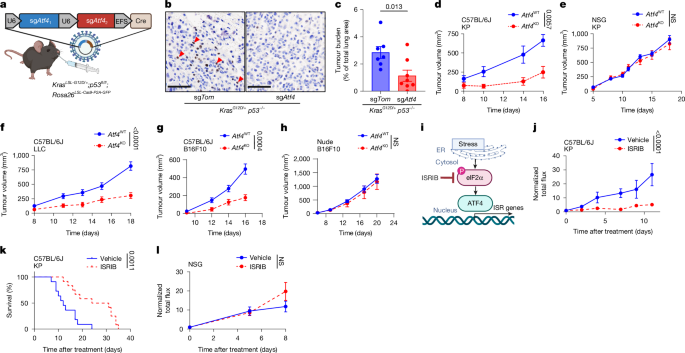
All experiments were approved by the New York University (NYU) Institutional Animal Care and Use Committee (IA16-01627). KrasLSL-G12D/+;Trp53fl/fl;Rosa26LSL-Cas9-P2A-GFP/LSL-Cas9-P2A-GFP (KPC) mice have already been described55,66,67,68,69,70,71. Mice aged six to eight weeks of both sexes with the appropriate genotype were selected to begin tumour initiation studies with the bifunctional lentiviruses (pUSEC), which express Cre recombinase and sgRNAs for conditional CRISPR–Cas9-based loss of Atf4 (sgAtf4) or control (sgTom). The sgRNAs used are listed in Supplementary Table 4. For the experiments with the Lcn2-loss GEMM, we used a double-guide lentiviral system with a control vector carrying sgNeo-1 and sgNeo-2, and KPC mice with Lcn2 loss infected with lentivirus carrying sgLcn2-1 and sgLcn2-2 guides. The total lung area occupied by each tumour was measured on haematoxylin and eosin (H&E)-stained slides using NIS-Elements software (Nikon). Transplant experiments were performed using nude (JAX strain 002019), NOD scid gamma (NSG; JAX strain 005557) or C57BL/6J (JAX strain 000664) mice. Cells (100,000 cells in 100 μl PBS) were injected s.c. into each flank of the mouse. B16F10 melanoma cells (2 × 105) were injected s.c. into female mice aged four to six weeks. LLC cells (5 × 105) were injected s.c. into female mice aged four to six weeks. Tumours were measured by caliper, and tumour volume was calculated according to the formula 0.5 × length × width2. Tumours were not allowed to grow more than 2 cm in any linear measurement, and did not exceed that limit in any of the experiments performed. To generate orthotopic lung tumours, KP cells expressing luciferase–GFP were injected intravenously (100,000 cells in 100 μl PBS) into male C57BL/6J (JAX strain 000664) mice, and tumour burden was measured by bioluminescence (Perkin Elmer IVIS Spectrum in vivo imaging system, d-luciferin, Perkin Elmer 122799). Data were analysed using Living Image software. Sample sizes are provided in the figure legends. The minimum sample size was predetermined on the basis of the stochastic variability expected in the individual model, and taking into consideration the difference in the expected effect magnitude. When possible, a higher number of mice was used to reduce waste and increase confidence. Precise numbers are provided in the figure legends.
For the PDAC orthotopic model experiments, 8- to 12-week-old female C57BL/6 mice were used. Syngeneic KPC7 cells, with or without Lcn2 knockout (Lcn2KO), were orthotopically implanted (200,000 cells per mouse) into the mice, and tumour growth was monitored weekly using ultrasound. Five weeks after implantation, the mice were euthanized, primary tumours were collected and the tissues were formalin-fixed for further IHC analysis.
In addition, KPC7 cells were orthotopically implanted (200,000 cells per mouse) into C57BL/6 mice. When the tumour size reached 100 mm3, the mice were randomized into two groups to receive either a control vehicle (Ctr) (n = 10) or anti-mLCN2 (n = 10) treatment by intraperitoneal (i.p.) injection. Pancreatic tumour burden was quantified by ultrasound imaging before and after the drug treatment. After the treatment, the mice were euthanized, and tumour tissues were collected for IHC studies. All experiments were approved by the NYU Institutional Animal Care and Use Committee (IA16-01627).
CRISPR screen using a library of sgRNAs targeting ATF4-controlled genes
More information on library cloning is provided in a previous report23. In brief, the oligo pool from a reconstituted oligonucleotide library synthesis (OLS) library was single-amplified with barcoded primers and inserted into an Esp3I-digested pUSEPB (U6-sgRNA-EFS-Puro-P2A-TagBFP) vector72. The screen included a total of 3,240 sgRNAs targeting 470 ATF4-controlled genes, 25 essential genes and 279 unique non-targeting Ctr sgRNAs (Supplementary Table 1). KP cells with stable expression of Cas9–blasticidin were generated by introducing the lentivector pLX_311-Cas9, which expresses blasticidin resistance from the SV40 promoter and Cas9 from the EF1a promoter, as described73. For the in vitro screen, at least 3.3 × 106 cells were maintained throughout the experiment to ensure a representation of at least 1,000×. A total of 6 × 107 cells were transduced with the sgRNA library in 10-cm dishes at a multiplicity of infection of 0.3, then selected with 10 μg ml−1 puromycin for 30 h. After this, the medium was refreshed for 24 h for medium without puromycin, after which cells were collected, counted and used for screening in vitro and in vivo. A fraction of cells representing 1,000× coverage were pelleted and stored at −80 °C (t0 population). Cells were passaged every 2–3 days, and 3.3 × 106 cells were plated after each passage. After 10 population doublings (t10), 3.3 × 106 cells were pelleted and stored at −80 °C. For the in vivo screen, the ATF4 CRISPR library was divided into two sub-libraries, containing 1,626 and 1,614 sgRNAs, respectively. KP cells were infected and selected in a similar manner as for the in vitro screen, and 1.6 × 106 were injected s.c. into the flanks of C57BL/6J (n = 4) and NSG (n = 4) mice. Twelve days after s.c. transplantation, tumours were resected and genomic DNA was isolated using lysis buffer (50 mM Tris, 50 mM EDTA, 1% SDS, pH 8.0) supplemented with 30 μl of 20 mg ml−1 proteinase K (QIAGEN, 19131) that was added to tissue (up to 200 mg) or cells (up to 30 million cells) and incubated at 60 °C overnight. This was followed by phenol:chloroform:isoamyl alcohol (25:24:1) phase separation, isopropanol precipitation and ethanol wash. Five micrograms of in vitro and 10 μg of in vivo samples’ genomic DNA were used for screen deconvolution with NEB Q5 High-Fidelity 2X Master Mix (M0492L). Integrated sgRNAs were amplified by PCR to attach sequencing adapters and barcodes, as described previously23. PCR products of around 248 bp in length were size-selected on 2% agarose and purified using the QIAquick Gel Extraction Kit (QIAGEN, 28704). Samples were sequenced on an Illumina NextSeq 500 (75-nt single-end reads) at NYU Genome Technology Center. sgRNA dropout results were identified using the MAGeCK algorithm74.
Cloning and virus generation
Cloning of CRISPR sgRNAs was performed as previously described into the USEC vector71. The list of sequences and primers is included in Supplementary Table 4. Lentivirus was generated by co-transfection of HEK293 cells with viral vector and packaging plasmids psPAX2 (Addgene, 12260) and pMD2.G (Addgene, 12259) using JetPrime transfection reagent (101000046). Medium containing the virus was collected 72 h after transfection and filtered through a 0.45-μm filter. For in vivo experiments, the virus was concentrated by ultracentrifugation at 25,000 rpm for two hours at 4 °C. The virus pellet was resuspended in PBS and stored at −80 °C until use. Virus was titred using the GreenGo reporter cell line71.
IHC
Sections were immunostained on a Leica BondRX automated stainer according to the manufacturer’s instructions. In brief, tissues underwent deparaffinization online, followed by epitope retrieval for 60 min at 100 °C with Leica Biosystems ER2 solution (pH 9, AR9640) and endogenous peroxidase activity blocking with H2O2 (provided in the Leica BOND Polymer Refine Detection System, DS9800). Sections were then incubated with primary antibodies against ATF4 (Cell Signaling Technology, 11815S, RRID: AB_2616025) at 1:50 for 60 min at room temperature. Primary antibodies were detected with anti-rabbit horseradish peroxidase (HRP)-conjugated polymer and 3,3′-diaminobenzidine (DAB) substrate that are provided in the Leica BOND Polymer Refine Detection System. After counter-staining with haematoxylin, slides were scanned at 40× on a Hamamatsu NanoZoomer (2.0-HT) and the image files were uploaded to the NYU Grossman School of Medicine’s OMERO Plus image data management system (Glencoe Software).
Chromogenic IHC for human LUAD biospecimens was performed on a Ventana Medical Systems Discovery Ultra platform using Ventana reagents and detection kits unless otherwise specified. Unconjugated polyclonal goat anti-human lipocalin 2 (LCN2, R&D Systems, AF1757, JBH0919031, RRID: AB_354974) antibody was optimized on a formalin-fixed, paraffin-embedded 14-core tissue microarray containing normal brain, liver and kidney. All samples were sectioned at 4 μm, collected onto plus microscope slides (Thermo Fisher Scientific, 22-042-924) and stored at room temperature before use. Initial optimization testing determined antigen retrieval requirements at a fixed concentration. Subsequent optimization manipulated the concentration and/or incubation to establish the final protocol parameters. In addition, unconjugated rabbit anti-human ready-to-use CD3, clone 2GV6 (IVD CD3, Ventana Medical Systems, 790-4341, J27879, RRID: AB_2335978) was assayed according to the manufacturer’s instructions. In brief, slides were heated at 60 °C for one hour and deparaffinized on-instrument using Discovery Wash (Ventana Medical Systems, 950-510) at 69 °C for 24 min. Antigen retrieval was performed in Cell Conditioner 1 (Ventana Medical Systems, 950-500) for 24 min for LCN2 and 64 min for CD3, both at 91 °C. Slides were treated with 3% H2O2 for 8 min to quench endogenous peroxidase. Anti-LCN2 was diluted 1:100 in Dulbecco’s PBS (Thermo Fisher Scientific, J67670.AP) and incubated for three hours at room temperature. Primary antibody was detected with rabbit anti-goat HRP-conjugated multimer (Ventana Medical Systems, 760-4311) and incubated for 8 min, followed by ChromoMap (760-159) DAB detection. CD3 was applied neat and incubated for 32 min at 37 °C, followed by ultraView DAB detection (Ventana Medical Systems, 760-500). Slides were washed in distilled water, counterstained with haematoxylin, dehydrated and mounted with permanent medium. Negative controls consisted of the primary antibody substituted with antibody diluent.
For multiplex immunofluorescence staining with Akoya Biosciences Opal reagents, slides were incubated with the first primary antibody and secondary polymer pair, and then underwent HRP-mediated tyramide signal amplification with a specific Opal fluorophore. The primary and secondary antibodies were subsequently removed with a heat retrieval step, leaving the Opal fluorophore covalently linked to the antigen. This sequence was repeated with subsequent primary and secondary antibody pairs and a different Opal fluorophore at each step (see Supplementary Table 5 for reagent details). Sections were counterstained with spectral DAPI (Akoya Biosciences, FP1490) and mounted with ProLong Gold Antifade (Thermo Fisher Scientific, P36935). Semi-automated image acquisition was performed either on a Leica AT2 whole-slide bright-field scanner at 40× magnification or on an Akoya Vectra Polaris (PhenoImager HT) multispectral imaging system at 20× magnification using PhenoImager HT 2.0 software in conjunction with Phenochart 2.0 and inForm 3.0 to generate unmixed whole-slide qptiff scans. Image files were uploaded to the NYU Grossman School of Medicine’s OMERO Plus image data management system (Glencoe Software).
Histology analysis
Tumour microarray samples from biopsies from patients with LUAD were assessed and graded as previously described52. Levels of LCN2 and TILs in individual tumour biopsies were graded in a blinded manner. PDAC human biospecimens from the tissue microarray panel were annotated and graded in a blinded manner, followed by quantification of CD3- and LCN2-positive cells within the tumour area. Only biopsies with PDAC histology were included in the analysis.
In vivo treatments and T cell depletion
Treatment with ISRIB (0.25 mg per kg) or vehicle control (1:1 dimethyl sulfoxide (DMSO), Thermo Fisher Scientific, D128-500, and polyethylene glycol 400) was administered daily through i.p. injections. For T cell-depletion experiments, anti-CD8 (2.43, BioXcell BE0061), anti-CD4 (YTS 191, BioXcell BE0119) or isotype control (rat IgG2b, LTF-2, BioXcell BE0090) was administered at 150 μg i.p. twice a week. For treatments with anti-PD1 (29F.1A12, BioXcell), antibodies were diluted in PBS and injected i.p. at 200 μg per mouse three times a week until the end point of the experiment.
ExCITE-seq
Mice were sedated with ketamine and xylazine, and were then injected with 2 μg of APC anti-CD45 (2 μg per mouse diluted in 100 μl PBS; BioLegend, 30-F11) retro-orbitally 3 min before lung collection. The lung lobes were minced on a glass slide and then digested (collagenase IV (Sigma-Aldrich, C5138) and DNAse I (Sigma-Aldrich, DN25) in RPMI with 10% FBS) for 35 min at 37 °C. Digestion was stopped with EDTA (1 mM). Digested tissue was filtered into a single-cell suspension through a 100-μm filter, followed by red blood cell lysis (BD Pharm Lyse, 555899). Cells were then washed and suspended in a staining buffer. Cells were then stained with live–dead staining (Zombie UV fixable viability dye; BioLegend, 423107) and PeCy7 anti-CD45 (see ‘Flow cytometry’ for staining protocol).
Approximately 500,000 lung immune cells from each mouse were sorted as live+IV−CD45−CD45+, and 50,000 tumour cells were sorted as live+CD45−GFP+. Sorted samples were multiplexed using cell hashing antibodies (BioLegend) and stained with antibody-derived tags. Cells from each sample were pooled and loaded into 10X Chromium75,76. Gene expression, together with hashtag oligo (HTO) libraries, was processed using Cell Ranger (v5.0.0) in multi-mode. Cell-containing droplets were selected using the HTODemux function available in the Seurat programme. Unique molecular identifier (UMI) count matrices from each modality were imported into the same Seurat object as separate assays. Viable cells were filtered on the basis of having more than 200 genes detected and less than 10% of total UMIs stemming from mitochondrial transcripts. HTO counts were normalized using centred log ratio transformation before hashed samples were demultiplexed using the Seurat::HTODemux function. Protein counts were normalized using the centred log ratio transformation. RNA counts were normalized using the Seurat::SCTransform function with regressions of cell-cycle score and ribosomal and mitochondrial percentages. Multimodal integration was performed using the weighted nearest neighbour method in Seurat. In brief, a weighted nearest neighbour network was constructed on the basis of modality weights estimated for each cell using the Seurat::FindMultiModalNeighbors function with the top 40 and top 30 principal components from normalized RNA and protein counts, respectively. A shared nearest neighbour graph was then built on the basis of the first 40 principal components, followed by identification of cell clusters using the Leiden algorithm and Seurat::FindClusters function at several resolutions to identify potential rare cell types. Cell types were annotated on the basis of canonical cell-type markers and differentially expressed genes of each cluster identified using the Seurat::FindAllMarkers function with a logistic regression model. Clusters expressing markers of the same cell type were merged into a single cluster. Cells were then projected onto a uniform manifold77 using the top 40 principal components for visualization. Data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE308689.
Flow cytometry
Mice were euthanized, and lungs were digested into a single-cell suspension as described above. Single cells were transferred to a 96-well round-bottom plate and resuspended in fluorescence-activated cell sorting (FACS) buffer (0.5% bovine serum albumin (BSA), 0.1% sodium azide and 1 mM EDTA). Live–dead staining was initially performed per protocol (Zombie UV fixable viability dye; BioLegend, 423107). Cells were then blocked with Fc block (2.4G2, BioXcell) for 10 min on ice. Antibody cocktail for surface staining was then added for 15 min on ice (see antibody list in Supplementary Table 5), and then the samples were washed with FACS buffer. Cells needing intracellular staining for FOXP3 were fixed and permeabilized using the FOXP3 staining buffer kit (eBioscience, 00552300). Intracellular Fc blocking was applied for 10 min on ice, followed by intracellularly staining with FOXP3 antibody for one hour on ice. Cells were then washed and resuspended in FACS buffer. Next, cells were washed, transferred to a 96-well round-bottom plate and resuspended in FACS buffer. Surface staining was done as described above. The cells were then washed and resuspended in FACS buffer. The samples were filtered with a 100-μm filter and then run on a BD LSRFortessa. Data were analysed using FlowJo v10.
LUMICKS AFS
Two hundred microlitres of 10.57-µm far-red melamine resin beads (Microparticles, MF-FluoRed-AR1286) were coated with LCN2 or F4/80 antibody. Unless specified, all volumes were 200 µl, incubations were done at room temperature on a platform rocker and two washes were performed with 1× PBS after each incubation. Sequentially, beads were incubated with 0.01% poly-l-lysine (Sigma-Aldrich, P4707) for 30 min, 100 mM BS(PEG)9 (Thermo Fisher Scientific, 21582) for 20 min, 0.5 mg ml−1 avidin (Thermo Fisher Scientific, 21128) for 20 min and 5 µM of biotinylated LCN2 or F4/80 antibody for 20 min. Beads were washed three times with 1× PBS, resuspended in 200 µl of 1× PBS and kept on ice until the avidity experiment.
BMDMs were disassociated from culture flasks and resuspended at a concentration of 8 × 107 cells per ml, and seeded on z-Movi (LUMICKS) microfluidic chips that were coated with poly-l-lysine (Sigma, P4707). Z-Movi chips seeded with BMDMs were placed in a 37 °C incubator for at least two hours for attachment, then 20 µl of protein or antibody on beads was flowed onto the z-Movi chip and incubated with BMDMs for 2 min. Anti-mouse or anti-human LCN2 Fabs (1 µM) were co-injected with 20 µl of protein or antibody on beads. After incubation, an acoustic force ramp from 0 to 1,000 pN over 2 min 30 s was applied within the z-Movi chip, and protein and antibody on-bead detachment was observed using real-time fluorescence imaging on the z-Movi system. Replicates were performed on different z-Movi chips with randomized run orders for protein and antibody conditions.
Avidity experiments were processed using proprietary Oceon software. Brightfield and fluorescence pre-flow images and time-lapse movies were loaded in Oceon. One hundred to two hundred events were recognized within the field of view for each run. Oceon software drew a region of interest at a single cell size. Detachment events were recognized as loss of fluorescent signal within the region of interest. Using pre-flow images, beads that remained stuck from previous runs were automatically removed from analysis. Beads that were bound to exposed glass and not target cells were also automatically excluded. Files produced by Oceon were imported into Microsoft Excel for organization before plotting with GraphPad Prism 10.2.3. Avidity data were plotted as (1) a function of force and (2) a bar chart at the indicated force.
Cultures of BMDMs, monocytes and activated T cells
Macrophages and pan-monocytes were derived from the tibiae and femurs of C57BL/6 male mice (age 6–10 weeks). Pan-monocytes were isolated from the bone-marrow cell suspension using a pan-monocyte isolation kit (Miltenyi Biotec, 130-100-629) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated FBS (HIFBS), 1% penicillin–streptomycin (Gibco, 15-140-122), 1% sodium pyruvate (Gibco, 11-360-070) and 0.05 mM 2-mercaptoethanol (Gibco, 21-985-023). Lcn2KO and Lcn2KO+mLcn2 cancer cells were cultured in RMPI supplemented with 10% HIFBS, 800 μg ml−1 hygromycin (Gibco, 10-687-010) and 50 μg ml−1 gentamicin (Gibco, 15-750-060).
Mouse spleens were collected to extract CD8a+ T cells using the CD8a+ T Cell Isolation Kit (Miltenyi Biotec, 130-104-075). T cells were activated by stimulation with IL-2 (BioLegend, 575402) and activation beads from the T Cell Activation/Expansion Kit (Miltenyi Biotec, 130-093-627) for three days and cultured in RMPI supplemented with 10% HIFBS, 1% penicillin–streptomycin, 1% sodium pyruvate, 1% non-essential amino acids solution (Gibco, 11-140-050), 1 M HEPES (Gibco, 15-630-106) and 0.05 mM 2-mercaptoethanol.
ChIP
KP cells were seeded at a density of 2.2 × 106 per 15-cm tissue culture plate and incubated for 30 h. Cells were treated with tunicamycin (200 ng ml−1) for 15 h without medium replacement; control samples received an equivalent volume of PBS. Cells were cross-linked in 1% formaldehyde in PBS for 8 min at 37 °C. Cross-linking was quenched by washing once with 10 ml of 5 mg ml−1 BSA in cold PBS (3 min, room temperature), followed by two washes with cold PBS. Cells were scraped in 700 μl cold PBS, pelleted (2,000 rpm, 2 min) and lysed in 1 ml of 1% SDS ChIP lysis buffer supplemented with 1× protease inhibitor cocktail (Thermo Fisher Scientific, P178444). Lysates were incubated on ice for 20 min and stored at −80 °C. Chromatin fragmentation was performed with sonication (Bioruptor Plus, high intensity, 21 cycles; 30 s on, 30 s off). DNA was purified using the Zymo-Spin ChIP kit following the manufacturer’s instructions: cross-link reversal at 65 °C for 90 min with proteinase K, followed by DNA binding, washes and elution in 8 μl DNA elution buffer. DNA integrity and fragment size distribution were confirmed by 1.5% agarose gel electrophoresis. Chromatin immunoprecipitation: protein G dynabeads (50 μl; Invitrogen, 10-003-D) were washed twice with PBS + 5 mg ml−1 BSA and incubated with 5 μg antibody (ATF4 or IgG control) for six hours at 4 °C. Antibody-coated beads were washed and incubated with 500 μl sonicated chromatin overnight at 4 °C. Beads were washed six times with RIPA buffer (with 15-min rotations every two washes) and twice with cold TE buffer. Elution and reverse cross-linking: beads were resuspended in 250 μl TE buffer with 5 μl RNase A (Qiagen, 19101) and incubated for one hour at 37 °C, followed by proteinase K digestion (1 μl, 15 min, 65 °C). Chromatin was eluted in 50 μl 2× elution buffer (Zymo) for 30 min at 65 °C with vortexing every 5 min, and reverse cross-linking was continued for one hour at 65 °C. DNA was purified with the Zymo-Spin ChIP kit and eluted in 20 μl elution buffer for downstream qPCR analysis.
siRNA knockdown in monocytes
Pan-monocytes were seeded under suspension culture conditions in 24-well flat-bottom ultra-low-adhesion plates at day 0 and transfected with 10 μM of Slc22a17 siRNA (mm.Ri.Slc22a17.13.1) or scrambled negative control siRNA (Qiagen, 102728) using lipofectamine RNAiMAX reagent (Invitrogen, 13-778-075) in OptiMEM medium (Gibco, 31985062) following the manufacturer’s protocol, in the presence of 5 ng ml−1 macrophage colony-stimulating factor (M-CSF) to promote cell survival. After 72 h, cells were collected and used for 3D monocyte infiltration.
Infiltration of monocytes in 3D matrices
Lcn2KO and Lcn2KO+mLcn2 cancer cells were seeded in a 2 mg ml−1 collagen type I (Thermo Fisher Scientific, CB-40236) solution at a concentration of 2 × 106 cells per ml, and placed in each well of a 96-well plate. These collagen solutions were allowed to polymerize for 30 min, forming a 3D matrix. Next, 50,000 pan-monocytes stained with CellTracker Deep Red (Invitrogen, C34565) were seeded in 200 µl of medium per well. For the blocking antibody experiment, the LCN2 blocking antibody was added at a concentration of 1 µg ml−1. For the siRNA knockdown experiment, pan-monocytes were seeded with penicillin–streptomycin-free DMEM. After 24 h of co-culture, the plate was imaged using the ImageXpress Micro Confocal system to assess the number of monocytes recruited inside the collagen matrix, using five z-stacks at a 20-µm interval. The number of monocytes was quantified using the TrackMate plug-in in ImageJ.
Infiltration of CD8 T cells in 3D matrices
Bone-marrow cells were seeded in a 96-well plate (2 × 105 cells per well) and were treated with 20 ng ml−1 M-CSF for five days to induce macrophage differentiation. Macrophages were co-cultured with Lcn2KO or Lcn2KO+mLcn2(Lcn2WT) cancer cells at a 100:1 ratio for two days in 96-well plates. Next, these cancer-conditioned macrophages (2 × 107 cells per ml) and cancer cells (2 × 107 cells per ml) were seeded in a collagen 3D matrix, followed by the seeding of 5 × 104 CellTracker Deep Red-stained CD8 T cells within each well. For the blocking antibody conditions, LCN2 and CXCL9 blocking antibodies were added at concentrations of 1 µg ml−1 and 40 µg ml−1, respectively. After 48 h of co-culture, the plate was imaged using a Zeiss LSM700 Confocal to assess the number of CD8 T cells recruited into the collagen matrix, using three z-stacks at a 15-µm interval. The number of CD8 T cells was quantified using the TrackMate plug-in in ImageJ.
Antibody discovery and characterization
Synthetic genes encoding human LCN2 (Uniprot ID: P80188) and mouse LCN2 (Uniprot ID: P11672), including their secretion signal sequences (residues 1–20), were cloned into the plasmid pBCAG in such a way that they were fused C-terminally with an Avi-tag and a polyhistidine tag. The plasmids were used to transfect Expi293F cells following the standard protocol from the vendor (Thermo Fisher Scientific). Transfected cells were incubated at 37 °C with 8% CO2 and collected on day 7 after transfection. After removing cells by centrifugation, the supernatant was dialysed against 20 mM sodium phosphate buffer pH 7.4 containing 500 mM NaCl. The dialysed solution was filtered and loaded onto a HisTrap excel column (Cytiva) pre-equilibrated in 20 mM sodium phosphate buffer pH 7.4 containing 500 mM NaCl, and eluted with 20 mM sodium phosphate buffer pH 7.4, containing 500 mM NaCl and 0.5 M imidazole. Eluted fractions from the HisTrap column containing the expressed protein were pooled and dialysed against 20 mM sodium phosphate buffer pH 7.4 containing 500 mM NaCl. For in vitro biotinylation, the protein was dialysed against 50 mM bicine buffer pH 8.0, and the biotinylation reaction was initiated by adding 10 mM magnesium acetate, 10 mM ATP, 0.5 mM biotin and in-house-prepared BirA enzyme at a 0.1 molar ratio to LCN2 (all final concentrations). The reaction mixture was incubated at 30 °C for one hour, and the LCN2 protein was purified with a gravity-flow Ni-NTA column and dialysed against gel filtration buffer (50 mM Tris HCl buffer pH 7.5 containing 150 mM NaCl). Finally, the sample was loaded onto a Superdex S75 Increase 10/300 GL size-exclusion column (Cytiva). Fractions containing the LCN2 protein were pooled, concentrated, aliquoted and stored at −80 °C until needed.
The sorting of a synthetic human antibody library was performed as described previously78,79. In brief, the phage library was incubated with biotinylated antigens at concentrations of 100 nM in the first and second rounds, 50 nM in the third round and 20 nM in the fourth round. Phage clones from the sorted library were assessed by phage multiplex bead binding assay80. The genes for Fab clones were subcloned into a bacterial expression vector using Golden Gate assembly (NEB)81, and Fab samples were produced from the 55244 Escherichia coli strain (ATCC) using published procedures78,79. Antibodies in the mouse IgG1 format were produced by Biointron.
Biolayer interferometry measurements were performed in 50 mM Tris HCl buffer pH 7.5 containing 150 mM NaCl, 0.5% BSA and 0.005% Tween 20 at 30 °C on an Octet RED96e instrument (Sartorius). Streptavidin sensors were used to immobilize a biotinylated analyte, and binding to a nonbiotinylated ligand was measured. Data were analysed with Octet data analysis software v12.0.2.59 (Sartorius).
Data acquisition and preprocessing
RNA sequencing (RNA-seq) data for five cancer cohorts from the publicly available TCGA programme—lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), non-small cell lung cancer (NSCLC; LUAD + LUSC), pancreatic adenocarcinoma (PDAC) and skin cutaneous melanoma (SKCM)—were downloaded via the UCSC Xena platform in HTSeq-FPKM format. Only tumour samples were retained.
Fragments per kilobase of transcript per million mapped reads (FPKM) values were converted to transcripts per million (TPM) using the formula: TPM = (FPKM/sum(FPKM)) × 106. Subsequently, log2(TPM + 1) transformation was applied to stabilize variance and facilitate visualization. All analyses were done using log2-transformed TPM values unless otherwise specified.
Gene signature definitions
An ATF4-downregulated gene signature was compiled from a publicly available dataset published by Igarashi et al.82. For each tumour sample, a signature score was computed as the mean log2(TPM + 1) expression of genes in the signature.
LCN2 expression was analysed independently and also used to stratify samples into tertiles. The low, medium and high LCN2 expression groups were defined using the 25th and 75th percentiles of log2 (TPM + 1) LCN2 expression within each cancer type.
Multiplex analysis of cytokines in BALF
We used Luminex xMAP technology to quantitatively and simultaneously detect 32 mouse cytokines, chemokines and growth factors. The multiplexing analysis was performed by Eve Technologies using the Luminex 200 system (DiaSorin) with Bio-Plex Manager software (Bio-Rad). Thirty-two markers were measured in the samples using the Eve Technologies Mouse Cytokine/Chemokine 32-Plex Discovery Assay Array (MD32) as per the manufacturer’s instructions for use (MILLIPLEX Mouse Cytokine/Chemokine Magnetic Bead Panel, MCYTOMAG-70K, MilliporeSigma). The 32-plex consisted of eotaxin/CCL11, G-CSF/CSF-3, GM-CSF, GROα/CXCL1/KC/CINC-1, GROβ/CXCL2/MIP-2/CINC-3, IFN-γ, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-12(p40), IL-12(p70), IL-13, IL-15, IL-17, IP-10/CXCL10, LIF, LIX, MCP-1/CCL2, M-CSF, MIG/CXCL9, MIP-1α/CCL3, MIP-1β/CCL4, RANTES/CCL5, TNF and VEGF-A.
Assay sensitivities of these markers range from 0.3 to 30.6 pg ml−1. Individual analyte sensitivity values are available in the MilliporeSigma MILLIPLEX protocol.
Correlation and stratified analysis
Pairwise Pearson correlation between LCN2 expression and the ATF4-downregulated gene signature was computed within each cancer cohort. For stratified analysis, samples were grouped by LCN2 expression tertiles. Groupwise differences were assessed using Wilcoxon rank-sum tests, and P ≤ 0.05 was considered statistically significant.
AI-based WSI analysis and gene-expression profiling of immune phenotypes in TCGA pan-cancer cohort
For spatial immune profiling and associated molecular marker analysis, we used transcriptomic data and WSIs from TCGA, encompassing 23 types of epithelial solid tumour: bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), oesophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumour (TGCT), thyroid carcinoma (THCA), uterine corpus endometrial carcinoma (UCEC) and uveal melanoma (UVM). All materials were accessed through the Genomic Data Commons Portal (https://portal.gdc.cancer.gov/).
For quantitative analysis of H&E WSIs, we used Lunit SCOPE IO, an artificial intelligence (AI) model developed using more than 26 tumour types. The cell detection model was constructed using 20,617 patches extracted from 5,609 WSIs (3,798 for training and 1,811 for optimization). The tissue segmentation component used 76,110 patches from 18,935 WSIs (15,936 for training and 2,999 for optimization). Board-certified pathologists annotated 2,828,448 cells, 1.70 × 1010 μm2 of cancer and stromal regions, 706,320 lymphocytes and 56,972 macrophages to train the model. The spatial distribution of TILs was evaluated by partitioning each WSI into 0.25-mm2 grids regardless of image dimensions. The AI system quantified both intratumoural and stromal TIL densities and categorized immune phenotypes on the basis of established thresholds: the inflamed phenotype required intratumoural TIL density ≥130 per mm2; the immune-excluded phenotype was defined by intratumoural TIL density <130 per mm2 with stromal TIL density ≥260 per mm2; and the immune-desert phenotype required TIL densities below these thresholds in both compartments. The predominant immune phenotype for each WSI was determined by grid distribution analysis: inflamed when ≥33.3% of grids displayed inflamed characteristics; immune-excluded when ≥33.3% of grids showed immune-excluded patterns while inflamed grids remained <33.3%; and immune-desert in all other cases.
The cellular density metrics were calculated as follows:
-
Intratumoural TIL density = lymphocyte count in cancer area/cancer area (mm2).
-
Stromal TIL density = lymphocyte count in cancer stroma/stromal area (mm2).
-
Macrophage density = macrophage count in cancer and stroma/combined area (mm2).
RNA expression data from TCGA, quantified as RNA-seq by expectation maximization (RSEM) values, underwent log2(RSEM + 1) transformation for scaling. We examined the differential expression of LCN2 and ATF4 between inflamed and immune-excluded phenotypes using analysis done with the Wilcoxon rank-sum test (Mann–Whitney U test), implemented in Python’s scipy.stats library to identify significant differences between these specific immune contexts. Statistical significance was determined at P < 0.05, and results were visualized using box plots with individual data points to represent the distribution of gene expression across these two phenotypes.
Furthermore, we stratified cases into three groups (low, moderate and high) based on LCN2 expression levels, and compared several microenvironmental variables, including stromal-to-intratumoural TIL ratio and macrophage density, using the same statistical approach.
AI-based WSI analysis and survival analysis of LCN2 expression in a validation cohort
The validation cohort consisted of 380 patients with NSCLC who were treated at Samsung Medical Center (SMC) with ICIs, including anti-PD1, anti-PD-L1 and anti-CTLA-4 agents, administered as either monotherapy or combination therapy, between September 2014 and July 2022. Immune phenotypes were determined using Lunit SCOPE IO following the same methodology as that described for the TCGA cohort. RNA expression data, quantified as TPM values, underwent log2(TPM + 1) transformation for scaling.
To investigate the association between LCN2 expression and immune exclusion patterns, we compared LCN2 expression levels between inflamed and immune-excluded phenotypes using the Wilcoxon rank-sum test. To elucidate the relationship between LCN2 expression and immune exclusion patterns, survival analysis was conducted exclusively on patients with inflamed and excluded immune phenotypes. Overall survival was defined as the time from treatment initiation to death from any cause or last follow-up. Patients were stratified into high and low LCN2 expression groups on the basis of the median expression value, and survival differences were evaluated using Kaplan–Meier analysis and Cox proportional hazards models.
Statistics
Statistical analysis was performed using GraphPad Prism v9. All data are expressed as mean and s.e.m., unless otherwise specified. Data were analysed by the statistical tests indicated in the figure legends. All tests were two-tailed unless specified otherwise.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.