Statistical model of fly brain
Here we approximate fly brain dynamics as a first-order vector auto-regressive model (VAR(1)),
$${{\bf{r}}}_{t}=W{{\bf{r}}}_{t-1}+{W}_{l,x}{L}_{t}+{{\epsilon }}_{t}.$$
(3)
where rt is the DâÃâ1 vector of responses from all neurons on time step t, W is the full DâÃâD dynamics matrix, Lt is the random DlâÃâ1 vector of stimulation power (for example, voltage), Wl,x is the matrix specifying the effect of stimulation, and ϵt is additive noise with arbitrary bounded covariance (for graphical model, see Extended Data Fig. 1). The raw IV estimator that we outline below requires no distributional assumptions on the random variables (Lt, ϵt). Furthermore, the noise (ϵt) can take on any correlation structure over time and across neurons, so our estimator applied to dynamics of this form will converge to the true effectome W, regardless of confounding inputs (so long as the noise is independent of the stimulation Lt). We note that for nonlinear dynamics, further assumptions are required (see Supplementary Information, âIV estimator applied to nonlinear dynamicsâ).
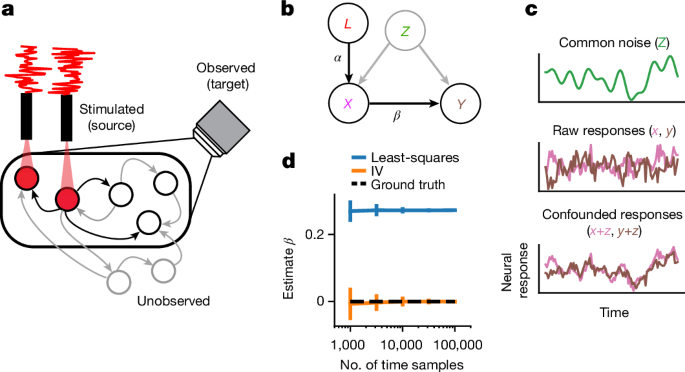
For our derivations below we define three subsets of neurons in rt, we will call the vector of neurons that are observed in a given experiment, target neurons, at time point t, Yt, the neurons that are both stimulated by the laser and observed Xt, source neurons, and finally neurons that are neither observed nor stimulated Zt (for graphical model, see Extended Data Fig. 1). Note that source neurons are a subset of target neurons.
Simulations
Simulations to analyse eigencircuits
To evaluate neural dynamics associated with eigencircuits (see âDominant circuits are interpretableâ) in continuous time, we restricted our analyses to neurons with the highest eigenvector loadings that accounted for 75% of all eigenvector power. In the case of the opponent motion circuit, this was 5 neurons, and in the case of the ellipsoid body circuit, this was 21 neurons. We set the discrete time step to Tâ=â10âms, and a sampling period of Îâ=â1âms so that at each time step,
$${{\bf{r}}}_{t+\varDelta }=\exp \left(\log \left(W\right)\varDelta {T}^{-1}\right)f\left({{\bf{r}}}_{t}+{{\bf{u}}}_{t}\right)$$
(4)
where W is the putative effectome of the subset of neurons being simulated, f(·) is a nonlinearity (here rectification), and ut is a vector of time-varying inputs.
We tuned the membrane time constant manually to 0.5âms to recapitulate the qualitative findings of prior work. The resulting W was scaled to have a maximum eigenvalue of 1. It is unknown what a good approximate scaling of synapse number to linear effect is in these regions but if the scaling was zero then this circuit would not perform a computation and if it was too large the dynamics would be unstable. The membrane time constant is often faster in sensory systems but depends on the voltage of the neuron42. Future work could consider other normative motivations for scaling weights and time constants (for example, ref. 32).
In both the ellipsoid and opponent motion simulations, inputs in Fig. 4 were simulated by adding in a step function input to the relevant neurons in our dynamics simulation. This step function was either at 0 or a hand tuned maximum. In the opponent motion circuit input maximum was 0.01 and for the ellipsoid body circuit the winner neuron receiving a larger input had input of 0.01 and other neurons had 0.006. In both cases stimulation lasted 150âms. In the case of the opponent motion circuit, we used prior literature to determine which neurons to stimulate under unilateral and bilateral BTF motion20,21. In the case of the ellipsoid body circuit, the ordering of the neurons with respect to retinotopic input was arbitrarily set by their eigenvector loading magnitude.
Simulations to evaluate estimators
In all simulations to evaluate statistical estimators, \({{\epsilon }}_{t} \sim {N}_{D}\left(0,{\varSigma }_{{\epsilon }}\right)\) where \({{\Sigma }}_{{\epsilon }}=c{I}_{D}\) and \({L}_{t} \sim {N}_{D}\left(0,l{I}_{D}\right)\). To simulate misspecification of the connectome prior mean, we estimated the accuracy of our estimator across many âground truthâ effectomes drawn from the connectome prior (except without a small constant added to the variance so that synaptic weights equal to 0 remained 0), such that the connectome prior mean was never the same as the âground truthâ effectome in a given simulation.
Given that there are a host of unknowns with respect to a real experimental setting (such as imaging SNR, strength of laser effect or duration of recordings), we hand tuned these parameters to give reasonable rates of convergence. In our whole-brain IV simulations, signal-to-noise ratio (SNR)â=â10 (laser power relative to noise power). We note that while we have simulated from a parameter regime in which our estimator converges rapidly, there are many parameter settings where convergence is slow. We note that conditions of high noise and little effect of the laser are particularly challenging. Slow timescales are also challenging because they effectively filter out most of the power of the white noise perturbation (but for extension to correlated IVs, see ref. 27). Yet, we show analytically that our estimator is consistent and thus will converge with enough samples.
For clarity, in our example simulation we chose a single neuron to stimulate and estimate its downstream synaptic weights (Fig. 3). This neuron was chosen because it had a larger than typical number of downstream contacts. It is straightforward to estimate downstream weights for multiple neurons simultaneously (equation (7)), and we demonstrate this both for IV and IVâBayes (Extended Data Figs. 4â6) and IV in a conductance-based model (Extended Data Figs. 3 and 4). We note that in our simulations, as the number of independent perturbations increases (nl), there is effectively more noise overall both through second-order effects and in estimating the effect on a downstream neuron that itself is being perturbed. This is another pressure to keep the number of stimulated neurons low and the strength of perturbation minimal, but depends on the particularity of connectivity.
IV estimator for an LDS
We note that
$$\text{Cov}\left[{X}_{t},{L}_{t}\right]={W}_{l,x}$$
(5)
because the stimulation is assumed to have identity covariance. Thus, by calculating the sample covariance between the laser and simultaneous activity in the stimulated neurons we can obtain an unbiased estimate of the linear weighting of laser drive on each neuron. Similarly, we can obtain an unbiased estimate of the linear effect of the laser on all target neurons at the next time step,
$$\text{Cov}\left[{Y}_{t+1},{L}_{t}\right]={W}_{x,y}{W}_{l,x},$$
(6)
where Wx,y is the submatrix of W with postsynaptic effects of Xt on Ytâ+â1. We can then use equation (5) to identify Wx,y with
$${W}_{x,y}={W}_{x,y}{W}_{l,x}{\left({W}_{l,x}\right)}^{+},$$
(7)
where (Wl,x)+ is a pseudo inverse because we have not specified the rank of L. Only if nLââ¥ânS is this a true inverse and Wl,x is invertible. An equivalent but more intuitive approach is termed two-stage least-squares (2SLS), where in the first stage Lt is regressed on Xt to give \({\hat{X}}_{t}={\hat{W}}_{l,x}{L}_{t}\) and then \({\hat{X}}_{t}\) is regressed on \({Y}_{t+1}\) to give \({\hat{Y}}_{t+1}={\hat{W}}_{x,y}\,{\hat{X}}_{t}\). The IV estimator can also be extended to higher order AR processes by conditioning the estimator on multiple past time steps27.
We note that multi-synaptic effects can be derived from the estimated monosynaptic effects with powers of the effectome matrix. For example, if we have the effectome matrix W and input r is an input vector of all zeros except for neuron i, then the nth order synaptic effect is exactly Wnr (for example, nâ=â1 gives direct synaptic effects, nâ=â2 gives effects through up to two synapses, and so on).
The connectome prior
We use the 2SLS approach to motivate a consistent estimator from a Bayesian perspective. In short, we perform classical Bayesian regression for the second stage of regression using the connectome as a prior on the weights Wx,y. To be consistent with the most typical setting of Bayesian regression we first work out the case of multiple source neurons and a single target neuron (that is, learning a set of weights in the same row of W). We assume the conditional distribution of the output given the input is:
$${y}_{t}{\rm{| }}{{\bf{x}}}_{t} \sim {\mathscr{N}}({{\bf{w}}}^{{\rm{T}}}{{\bf{x}}}_{t},{\sigma }^{2}),$$
(8)
where \(\left({{\bf{x}}}_{t},{{\bf{y}}}_{t}\right)\) represents the input and output for sample \(t\in \{1,\ldots ,T\}\), and \({\sigma }^{2}\) is the variance of the observation noise in y.
Let us now suppose that μ provides the mean for a Gaussian prior over the linear weights w:
$${\bf{w}} \sim {\mathscr{N}}(\mu ,{\gamma }^{2}I).$$
(9)
Let μâ=âsc, where the hyperparameter s scales c, the connectome prior we set to be equal to the synaptic count and sign. Combining this prior with the likelihood defined above gives us the following posterior mean:
$$\begin{array}{l}{\hat{{\bf{w}}}}_{{\rm{M}}{\rm{A}}{\rm{P}}}={\text{arg}\text{max}}_{{\bf{w}}}P({\bf{w}}|X,Y,\theta )\\ \,\,\,=\,{(1/{\sigma }^{2}{X}^{{\rm{T}}}X+1/{\gamma }^{2}I)}^{-1}(1/{\sigma }^{2}{X}^{{\rm{T}}}Y+1/{\gamma }^{2}\mu )\\ \,\,\,=\,{({X}^{{\rm{T}}}X+{\sigma }^{2}/{\gamma }^{2}I)}^{-1}{X}^{{\rm{T}}}Y+{({\gamma }^{2}/{\sigma }^{2}{X}^{{\rm{T}}}X+I)}^{-1}\mu ,\end{array}$$
(10)
where \(\theta =\{{\sigma }^{2},{\gamma }^{2},c\}\) denotes the hyperparameters. The second expression above (equation (10)) shows that the maximum a posteriori (MAP) estimate is the standard âridge regressionâ estimate, \({\hat{{\bf{w}}}}_{{\rm{r}}{\rm{i}}{\rm{d}}{\rm{g}}{\rm{e}}}={\left({X}^{{\rm{T}}}X+\frac{{\sigma }^{2}}{{\gamma }^{2}}I\right)}^{-1}{X}^{{\rm{T}}}Y\), plus a term that biases the estimate towards the anatomical connectome μ. Note that in the limit of small observation noise \({\sigma }^{2}\) or large prior variance \({\gamma }^{2}\), the MAP estimate converges to the maximum likelihood (ML) estimate, whereas in the limit of large \({\sigma }^{2}\) or small \({\gamma }^{2}\), it converges to μ.
In our simulations we choose the optimal hyperparameters beforehand but the hyperparameters could be learned via a standard cross-validation grid search. A more principled approach would be to use evidence optimization,
$$\hat{\theta }=\text{arg}\,{\text{max}}_{\theta }P(Y{\rm{| }}X,\theta )=\text{arg}\,\mathop{\text{max}}\limits_{\theta }\int P(Y{\rm{| }}X,{\bf{w}},\theta )P({\bf{w}}{\rm{| }}\theta )d{\bf{w}},$$
(11)
which would be straightforward given that the evidence is available in closed form for this model.
Construction of fly connectome matrix
The connectome is a reconstruction of the central nervous system of a seven-day-old adult female Drosophilia melanogaster. We use the most recent version of the connectome v783. Details of the reconstruction are provided in the original publications of the connectome dataset1.
Each entry in the connectome matrix W, the main object of study in our analyses, was the number of synapses multiplied by their inferred sign based on predicted neurotransmitter type11. Specifically, neurons with neurotransmitters acetylcholine and dopamine had positive weights on their downstream neurons and neurons with GABA, serotonin, glutamate and octopamine had negative weights. The neurotransmitter type was predicted directly from electron microscopy images trained on synapses with known neurotransmitter types. The matrix W scaled for stability was used as the connectome prior mean in estimator simulations (Fig. 2) and our eigendecomposition analysis (equation (2); Figs. 3 and 4). A threshold was set on the synapse count such that any connections with less than five synapses were set to zero. This choice followed the reasoning of other analyses of the connectome10 that this would minimize the impact of spurious synapsesâmanual proofreading did not extend to connections with fewer than five synapses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.