Incorporation of small molecules in MaSIF
Molecular surface meshes were triangulated using the MSMS program61, and radial patches (geodesic radius 12 Å) were computed following the original MaSIF preprocessing scripts8. Before MaSIF’s geodesic convolutional layers are applied, five input features are computed for each patch: shape index31, distance-dependent curvature32, Poisson–Boltzmann continuum electrostatics, hydrogen bond donor and acceptor potential33, and hydropathy34,35,36. The first two features are purely geometric and are calculated analogously to protein surfaces alone. Moreover, the APBS program62 used to compute the Poisson–Boltzmann electrostatics on the surface supports small molecules in the MOL2 file format and hence does not require us to treat them in a conceptually different way. The remaining two chemical input features are computed as described below.
Hydrogen bond donors and acceptors
The hydrogen bond propensity feature assigns a positive value to points on the molecular surface near the optimal direction in which a hydrogen bond could be formed with an acceptor atom. It is determined by the direction of the covalent bond between a donor atom and its hydrogen (Supplementary Fig. 1b,c). Likewise, a negative value is assigned to points corresponding to hydrogen bond acceptors. For different acceptor types, the theoretically optimal position for forming a hydrogen bond can either lie on a cone (Supplementary Fig. 1d–f) or in a small number of specific directions that can be derived from the molecular geometry. We assign different magnitudes of the donor or acceptor feature on the basis of the angular deviation from the ideal hydrogen bond geometry according to a quadratic function.
The optimal direction of the hydrogen bond was determined using the RDKit software package63, and surface points were assigned positive (donor) or negative (acceptor) values between −1 and +1 on the basis of their angular deviation from the ideal direction. For potential acceptors, RDKit was also used to determine whether the idealized location of the hydrogen bond lay on a cone or in one or more discrete directions.
Hydropathy
MaSIF’s hydrophobicity feature makes use of the Kyte–Doolittle scale34, which is exclusively defined for amino acids. Equivalent values for small molecules thus need to be approximated on the basis of a more general hydrophobicity measure that can be estimated computationally, such as the logarithm of the octanol–water partition coefficient (logP)35. To this end, we developed a nonlinear function that maps logP values to the Kyte–Doolittle scale. We fit the parameters of this function to find an optimal match for the Kyte–Doolittle and logP values of all 20 amino acids. As the best functional form of this mapping was not immediately obvious from the raw values (Supplementary Fig. 1l), we experimented with different hydrophobicity scales as intermediates and found that the Eisenberg scale36 had approximately linear and exponential relationships with logP and Kyte–Doolittle values of amino acids, respectively. We first computed the optimal parameters of the mappings from logP to Eisenberg scale (Supplementary Fig. 1g) and Eisenberg scale to Kyte–Doolittle scale (Supplementary Fig. 1h) and then composed these two functions to establish the desired relationship between logP and Kyte–Doolittle values (Supplementary Fig. 1i). Finally, we also restricted the outputs to the valid interval of Kyte–Doolittle values [−4.5, 4.5] to ensure that the feature did not leave the domain on which MaSIF was trained.
Furthermore, as some ligands can cover large surface patches, we aimed to capture local variations of the hydrophobicity by fragmenting the molecules before calculating their hydrophobicity scores. We used the BRICS algorithm64 to decompose molecules and compute estimates of the logP value of each fragment with RDKit. The resulting fragments were more similar in size to amino acids and tended to have less extreme hydrophobicity scores than whole ligands, moving the distribution of this feature closer to that expected on protein surfaces (Supplementary Fig. 1k–l). To translate from logP to the Kyte–Doolittle scale, we parameterized a function so that it approximated the relationship between these hydrophobicity values for the 20 amino acids. Kyte–Doolittle and Eisenberg values of all amino acids are available in tabular form, whereas we computed their logP with RDKit to fit the curves. The final function was:
$$\beginarrayc\rmKyte-\rmDoolittle=\rmclip\,(-\,6.2786+\exp (0.4772\times \rmlogP+1.8491),\,\\ \,\,\,\,\,\,\,\,\min =-4.5,\,\max =4.5).\endarray$$
After computing equivalent Kyte–Doolittle values for all small-molecule fragments, we assigned the resulting hydrophobicity score of the closest fragment to each surface vertex.
To create the histograms in Supplementary Fig. 1k–l, we extracted 20,363 unique small-molecule ligands from the Binding MOAD65 dataset, fragmented each and removed duplicates. This resulted in 9,362 unique fragments that were compared with the set of ligands and the 20 standard amino acids.
Target protein selection
The target proteins were selected on the basis of several factors including the reported protein–ligand affinity66, the resolution of the structural data, the interface propensity, and the solvent-accessible surface area of the small molecule when bound to the receptor to ensure a measurable interface with the designed binders. More practical considerations such as small molecule purchase availability or feasibility of target protein expression were also considered.
Binding site identification
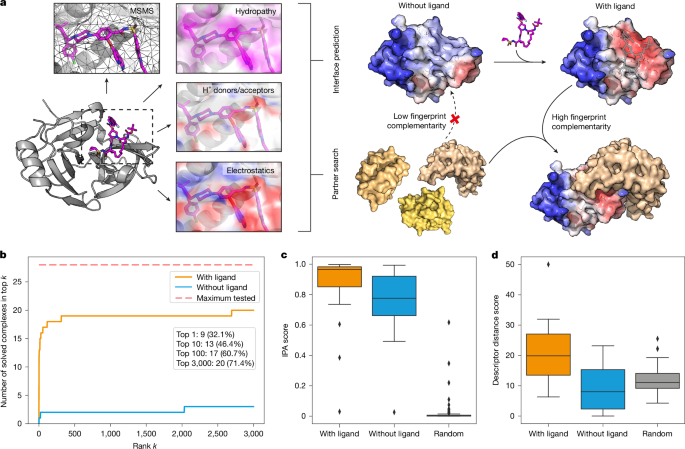
MaSIF-site8 was trained on a dataset of known PPIs to predict regions on protein surfaces with high propensity to form a buried interface. The neural network takes a protein–ligand complex decomposed into 12-Å (geodesic radius) overlapping patches as input and generates a per-vertex regression score, indicating the propensity of each point to become a buried surface area within a protein interaction. In this study, we used MaSIF-site to predict interfaces and guide the selection of target patches both in our computational benchmark and for all three target complexes for design (Bcl2–venetoclax, DB3–progesterone and PDF1–actinonin). In the computational benchmark, we conducted the search only for the three patches with the highest interface propensity near the centre of the binding site. For design, the number of targeted sites overlapping with the protein–ligand neosurface depended on the solvent-accessible surface area of each ligand to ensure that all the ligand-exposed surface was covered during the complementary motif search. This number was 1 for PDF1–actinonin, 2 for DB3–progesterone and 3 for Bcl2–venetoclax.
Binding seed identification
The fingerprints of the predicted 12-Å (geodesic radius) patches comprising both protein target and bound small molecule were used to find a complementary fingerprint in the MaSIF-seed database9, which contains approximately 640,000 continuous structural fragments (seeds) amounting to 402 million surface patches (also known as fingerprints). The seed database covers distinct secondary structures with approximately 390,000 sheet-based and 250,000 helical motifs. The MaSIF-search algorithm was trained to make patch fingerprints similar for interacting patches and dissimilar for non-interacting patches. Seeds with interface propensity scores above the defined threshold and with fingerprint distances (Euclidean distance between target and seed fingerprint) below the defined thresholds were selected. In second-stage alignment and scoring using the RANSAC algorithm, seeds were selected on the basis of IPA score. Cutoffs used for the seed selection are summarized in Supplementary Table 1.
Scoring aligned structures
We consider two descriptor-based postalignment scores. The descriptor distance score is a simple heuristic that aggregates descriptor distances across the predicted binding interface and is based on the squared Euclidean distances between interacting patches on each side of the interface. Two patches are considered to interact with each other if their centre points are less than 1.5 Å apart. The descriptor distance score is computed according to the following formula:
$$\rmD\rmD\rmS=\sum _i\frac1^2$$
where DDS is the descriptor distance score, i indexes interacting patches of the first protein and NN(i) returns the index of the spatially nearest neighbour on the other protein. Higher scores mean higher complementarity.
The IPA score is computed by a neural network that was trained to discriminate between near-native and high-r.m.s.d. poses of docked proteins8. The inputs of this predictor are three-dimensional Euclidean distances, descriptor distances and dot products between surface normals of up to 200 pairs of corresponding patches at the predicted interface. The predictor outputs values between 0 and 1, where larger values indicate higher confidence in the presented interface.
Computational binder recovery benchmark
The binder recovery experiment was performed for 14 known ligand-induced protein complexes, in which both proteins involved in the interaction are considered as separate items, resulting in 28 search queries. In addition, we included 8,907 decoys based on 2,852 PPIs from the PDBbind (v.2020)66 database. We split the provided structures into separate chains and only applied light filtering to remove nuclear magnetic resonance (NMR) structures, duplicate sequences within the same structure, and structures that could not be processed. All benchmark complexes and decoys are listed in Supplementary Table 4 and in the GitHub repository respectively (‘Code availability’). After triangulating and featurizing all protein surfaces with and without ligands, we screened the database and docked candidates, analogous to the binding seed search. Here we assumed the location of the binding site on the target protein was known and selected the three surface vertices with the largest predicted surface propensity within 10 Å of the centre of this site as input patches. The centre of the binding site was approximated with a simple heuristic. We first identified interface atoms as those within 4 Å of any atom from the binding protein in the original complex structure. This could and typically did include atoms belonging to the small molecule. Then, we defined the average of the coordinates of all interface atoms of the target protein as the centre of the binding site. Furthermore, we declared a binder to be correctly recovered if its i.r.m.s.d. compared with the ground truth structure of the same protein was less than 5 Å, where i.r.m.s.d. considered only heavy atoms in the immediate vicinity of the target protein (less than 5 Å).
Seed and interface refinement
To optimize binding energy of the seed for the target complex, seeds were refined using a FastDesign protocol on Rosetta37 with a penalty for buried unsatisfied polar atoms in the scoring function67. Refined seeds were then selected on the basis of the computed binding energy (ddG), shape complementarity, number of interface hydrogen bonds, number of buried unsatisfied polar atoms and number of atoms in contact with the small molecule. β-sheet-based motifs making more than 33% contact with the target complex using loop regions were discarded. Moreover, the uniqueness of each seed was assessed by a pairwise alignment of the hotspot residues. For seeds showing more than 70% hotspot identity with another seed, only the one with the best surface-normalized ddG was kept.
Seed grafting and computational design
For each target, approximately 100–120 selected seeds were subsequently grafted with a Rosetta MotifGraft68 protocol to stabilize the binding motif and bring further contacts with the target complex. Each seed was matched with a database of around 6,500 small protein scaffolds (less than 90 amino acids) originating from small globular monomeric proteins from the PDB69 and four computationally designed miniprotein databases that had been experimentally validated70,71,72,73. Before grafting on multiple scaffolds, seeds were cropped to the minimum number of residues making contact with the target, and loop motifs were removed from β-sheet-based seeds to optimize the grafting success rate. Once grafting had been performed, scaffolds underwent sequence optimization using a FastDesign protocol on Rosetta with a penalty for buried unsatisfied polar atoms in the scoring function. Final designs were selected based on the ddG, shape complementarity, number of interface hydrogen bonds and count of buried unsatisfied polar atoms. A similar number of designs per seed was ensured by setting dynamic cutoffs of these metrics adjusted for each seed.
Design optimization with LigandMPNN
Designs that did not show any binding in the first round of experimental screening underwent sequence optimization with LigandMPNN51. Ten sequences per design were generated and folded with AlphaFold2 in the ColabFold software50 (single sequence mode). Cα-r.m.s.d. values between AlphaFold2 predictions and the original model were measured, and only one sequence per design with the lowest r.m.s.d. was selected. Designs in complex with their respective target were relaxed with Rosetta and filtered based on the ddG, shape complementarity, number of interface hydrogen bonds and number of buried unsatisfied polar atoms. Five-hundred designs per target complex were selected and rescreened by yeast display.
Library screening
For each target complex, around 2,000 protein designs were reverse-translated into DNA and purchased from Twist Bioscience as oligo pools with 18-bp homology overhangs. Oligo pools underwent two rounds of PCR: (1) for amplification of the library using the 18-bp overhangs; and (2) for addition of 45-bp homology with the yeast display vector (57.5 °C annealing for 30 s, 72 °C extension time for 1 min, 15 cycles). EBY-100 yeast was transformed by electroporation using the amplified inserts and linearized HA-tagged pCTcon2 vector as described previously38. A similar approach was used for the SSM library of single designs. Transformed yeast cells were grown in minimal glucose medium (SDCAA) at 30 °C and induced with minimal galactose medium (SGCAA) overnight before sorting.
Yeast surface display of single designs
Genes encoding single designs were purchased from Twist Bioscience with an approximately 25-bp homology overhang for cloning. Each design was cloned into an HA-tagged pCTcon2 plasmid using Gibson assembly and transformed into XL10-Gold or HB101 bacteria for DNA production. The purified and sequence-approved DNA was then used to transform competent EBY-100 yeast using a Frozen-EZ Yeast Transformation II Kit (Zymo Research). For libraries, transformed yeast cells were grown in minimal glucose medium (SDCAA) at 30 °C and induced with minimal galactose medium (SGCAA) overnight before flow cytometry analysis.
Flow cytometry analysis and sorting
Induced yeast cells were washed with phosphate-buffered saline (PBS) supplemented with 0.1% bovine serum albumin and then labelled with the respective binding target for 2 h at 4 °C. Before labelling, protein–ligand complexes were preincubated at room temperature for 5 min with a 1:5–10 ratio. Cells were then washed and labelled with an FITC-conjugated goat anti-HA tag antibody (Bethyl, A190-138F; display tag; 1:100 dilution) and a PE-conjugated goat anti-human Fc antibody (Invitrogen, 12-4317-87; binding tag; 1:100 dilution) for 30 min at 4 °C. Cells were washed, resuspended in an appropriate volume of buffer and analysed on a Gallios flow cytometer (Beckman Coulter) or sorted with a Sony SH800 cell sorter. Kaluza software (Beckman Coulter, v.1.1.20388.18228) and LE-SH800SZFCPL Cell Sorter (Sony, v.2.1.5) were respectively used for the data acquisition. In the case of cell sorting, each designed library was sorted for binding and non-binding populations separately. Flow cytometry data were then analysed using FlowJo (BD Biosciences, v.10.8.1).
Library sequencing
Sorted yeasts were cultured and plasmids encoding protein designs were extracted using a Zymoprep Yeast Plasmid Miniprep II (Zymo Research) following the manufacturer’s protocol. The sequence of interest was then amplified by PCR with vector-specific primers flanking the protein design gene. A second PCR was performed to add Illumina adaptors and Nextera barcodes, and the PCR product was desalted and purified using a Qiaquick PCR purification kit (Qiagen). An Illumina MiSeq system with 500 cycles was used for next-generation sequencing. Around 0.8–1.2 millions reads per sample were obtained; these were translated into the appropriate reading frame and matched with expected input sequences from the libraries. The enrichment of each design was calculated by normalizing the counts in the binding population with the counts in the non-binding populations. Hits were identified if the enrichment was more than ten-fold and the number of counts in the binding population was greater than 10,000.
Protein expression and purification
A list of protein sequences can be found in Supplementary Table 5. Genes encoding the 6xHis-tagged and/or human Fc-tagged protein of interest were purchased from Twist Bioscience, cloned into pET11 (bacterial vector) or pHLSec (mammalian vector) by Gibson assembly and transformed into XL10-Gold or HB101 bacteria. Plasmids were extracted using a GeneJET plasmid Miniprep kit (Thermo Fisher, for bacterial vector) or a PureLink Fast Low-Endotoxin Midi plasmid purification kit (Invitrogen, for mammalian vector) and checked by Sanger sequencing. Proteins were purified using bacterial or mammalian expression systems. Mammalian expression was performed using an Expi293 expression system (Thermo Fisher, A14635). Cells were authenticated (short tandem repeat (STR) genotyping) and tested negative for mycoplasma contamination (quantitative PCR) by the provider. Supernatants were collected after 6 days and filtered and purified as described below. For bacterial expression, BL21(DE3) or T7 Express Competent Escherichia coli were transformed with the plasmid of interest and grown as a preculture overnight. Precultures were inoculated 1:50 in Terrific Broth medium and incubated at 37 °C until they reached an optical density at 600 nm (OD600) of approximately 0.7. Then, bacteria were induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and incubated overnight at 18–20 °C. Cells were collected by centrifugation at 4,000g for 10 min, resuspended in lysis buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 5% glycerol, 1 mg ml−1 lysozyme, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 µg ml−1 DNase) and lysed by sonication. Lysates were then clarified by centrifugation at 30,000g for 30 min and filtered.
All 6xHis-tagged proteins were purified using an ÄKTA Pure system (GE Healthcare) Ni-NTA HisTrap affinity column, followed by size-exclusion chromatography on a Superdex HiLoad 16/600 75 pg or 200 pg depending on the size of the protein. All proteins were concentrated in PBS as a final buffer.
Surface plasmon resonance
Affinity measurements were performed on a Biacore 8K (GE Healthcare, software v.4.0.8.19879) using HBS-EP+ as a running buffer (10 mM HEPES at pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% v/v surfactant P20; GE Healthcare). All proteins were immobilized on a CM5 chip (GE Healthcare, catalogue no. 29104988) by means of amine coupling to reach 500–1,000 response units. Analytes were then injected in serial dilutions using the running buffer. The flow rate was 30 μl min−1 for a contact time of 120 s, followed by 400 s of dissociation time. Surface plasmon resonance data were fitted in steady-state affinity mode by reporting the relative response units for each concentration.
Biolayer interferometry
Biolayer interferometry measurements were performed on a Gator system using GatorOne software (Gator Bio, v.2.7.3.0728). The running buffer consisted of 500 mM NaCl and 50 mM Tris pH 7.5 or HPS-P+ buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 1 μM NiSO4, 0.005% v/v surfactant P20; GE Healthcare) supplemented with 100 nM venetoclax or 5 μM actinonin if needed. Fc-tagged proteins were immobilized at a concentration of 7 μg ml−1 on protein A probes (1.5 to 2.5 nm immobilized) and dipped into serial dilutions of the ligand. Steady-state responses were normalized with the maximum value and plotted using a nonlinear four-parameter curve-fitting analysis.
Grating-coupled interferometry
Grating-coupled interferometry measurements were performed on a Creoptix WAVE system (Malvern Panalytical) using Creoptix WAVE control software (Malvern Panalytical, v.4.5.18). The running buffer consisted of HPS-P+ buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 0.005% v/v surfactant P20; GE Healthcare). All protein targets were immobilized on a 4PCH chip (Malvern Panalytical) by means of amine coupling to reach 7,000–10,000 pg mm−2. An intermediate injection with 1 μM NiSO4 was used for PDF1 protein. S55746, OBz-Pro and TBDMS-Act were then injected sequentially as analytes at concentrations of 2, 2.5 and 5 μM, respectively, using the waveRAPID (repeated analyte pulses of increasing duration) kinetic assay74. The flow rate was 100 μl min−1 for an injection duration of 25 s followed by 300 s of dissociation time for TBDMS-Act, whereas an injection duration of 50 s followed by 600 s of dissociation time was used for S55746 and OBz-Pro. Measurements were fitted with either a 1:1 model (for Bcl2–S55746 and PDF1–TBDMS-Act) or with a mass transport model (for BD3–OBz-Pro).
Size-exclusion chromatography combined with multiangle light scattering
Size-exclusion chromatography combined with multiangle light scattering (miniDAWN TREOS, Wyatt) was performed to determine the molecular weights of the purified designs. The final concentration was approximately 1 mg ml−1 in PBS (pH 7.4), and 100 μl of the sample was injected into a Superdex 75 10/300 GL column (GE Healthcare) with a flow rate of 0.5 ml min−1. Ultraviolet absorbance at 280 nm, differential refractive index and light scattering signals were recorded. Molecular weight was determined using ASTRA software (v.6.1, Wyatt).
Circular dichroism
Far-ultraviolet circular dichroism spectra were obtained with a Chirascan spectrometer (AppliedPhotophysics). Protein samples were diluted in PBS at a protein concentration of 300 μg ml−1 and placed in 1-m path-length cuvettes. Wavelengths between 200 nm and 250 nm were recorded with a scanning speed of 20 nm min−1 and a response time of 0.125 s. All spectra were corrected for buffer absorption. Temperature ramping melts were performed from 20 to 90 °C with an increment of 2 °C min−1. Thermal denaturation curves were plotted by the change in ellipticity at the global curve minimum. If possible, melting temperatures were determined after fitting the data with a sigmoid curve equation in GraphPad Prism.
Cell transfection and induction
Human embryonic kidney cells (HEK293T; Invitrogen, R70007) were cultured in Dulbecco’s modified Eagle medium (DMEM; 41966-029, Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS; A5256701, Gibco) and 1% (v/v) penicillin–streptomycin (15140-122, Gibco). Cells were authenticated by the provider (STR genotyping) and tested negative for mycoplasma contamination (quantitative PCR). Cells were maintained at 37 °C with 5% CO2 and passaged every 2–3 days at around 80% confluence. Cells were seeded into the inner 60 wells of a 96-well plate at 10,000 cells per well 24 h before to transfection. Cells were transfected by layering 50 μl from a mixture of 330 μl DMEM, 825–850 ng total DNA and 4.125 μg polyethylenimine (24765-1, Polysciences) on top of the medium in each well, enough for each six-well column with a 10% extra margin, as described previously75. Cells were left to incubate overnight, for a minimum of 12 h. The next morning, medium was replaced with fresh medium including the respective dilutions of the inducing agent.
Cellular detection assay
In the secreted NanoLuc assays, cells were seeded into clear 96-well cell culture plates (655-180, Greiner Bio-One) and transfected the next day. In the venetoclax-induced GEMS assay, cells were transfected with STAT3 (100 ng), STAT3-NanoLuc reporter (150 ng), and either a single GEMS receptor chain containing Bcl2 or DBVen1619_2 (600 ng) or both chains together (300 ng each). In the secreted split NanoLuc progesterone-induced assay (Extended Data Fig. 4), cells were transfected with 412.5 ng of scFv-DB3(VL/VH)-N-term-NanoLuc and 412.5 ng of DBPro1156_2-C-term-NLuc or 825 ng of a single plasmid. Cells were induced with their respective agent the following day. After 24 h of induction, 5 μl medium was transferred to a black 384-well plate (3820, Corning) and mixed with 5 μl diluted substrate from the Nano-Glo Luciferase Assay kit (N1120, Promega). After gentle shaking, plates were measured on a Tecan Spark plate reader with an integration time of 1,000 ms.
For intracellular NanoLuc assays, cells were plated in black 96-well cell culture plates (655086, Greiner). The next day, cells were transfected with either a single chain of PDF1-C-term-NanoLuc or DBAct553_1-N-term-NanoLuc (825 ng) or both chains together (412.5 ng each). The following day, cells were induced with different dilutions of the inducing agent actinonin. After 24 h of induction, intracellular nanoluciferase activity was measured using a Nano-Glo Live Cell Assay kit (N2012, Promega). Medium was aspirated and replaced with 24 μl RPMI medium (52400-025, Gibco) containing 10% v/v FBS, and 6 µl diluted substrate was added to each well. After gentle shaking, plates were measured on a Tecan Spark plate reader with an integration time of 1,000 ms. All cell-based fits presented in Fig. 5 and Extended Data Fig. 4 were calculated from technical replicates (n = 3) using a nonlinear four-parameter curve-fitting analysis. All statistical analyses were based on two-way analysis of variance (ANOVA) with multiple comparisons.
Cell-free reporter system
The gene encoding the 6xHis-DBPro1156_2 protein fused to T7 RNA polymerase (T7RNAP) was cloned into a pQE30 plasmid using Gibson assembly. The plasmid was then transformed into NEBExpress Iq competent E. coli (NEB, C3037I) for protein expression. Bacteria were precultured overnight and inoculated to a 500 ml Luria-Bertani (LB)-medium culture, grown until the OD600 was approximately 0.7 and then induced with 0.1 mM IPTG for 3 h. The cells were collected by centrifugation at 4,000g and lysed by sonication. Proteins were purified using Ni-NTA IMAC Sepharose gravity columns.
The ZF438-DB3 scFv (VH/VL) fusion protein was expressed using a PURExpress kit from NEB (E6800S) with the addition of a disulfide bond enhancer (E6820S). The reaction volume was 10 µl, containing 4 µl of solution A, 3 µl of solution B, 0.4 µl of NEB disulfide bond enhancer 1, 0.4 µl of NEB disulfide bond enhancer 2, 2 µl of DNA template (10 ng µl−1) and 0.2 µl of water. The reaction was incubated at 34 °C for 3 h and used for the following reporter reaction.
A PURExpress kit from NEB (E6800S) with disulfide bond enhancer (E6820S) was used to set up the mCherry reporter expression as well. The reporter-expressing reaction also included 100 nM purified DBPro1156_2-T7RNAP and ZF438-DB3 scFv pre-expressed with PURExpress. The DNA template for the mCherry gene was set to 4 nM, and the mCherry gene was transcribed under the regulation of a truncated T7 promoter downstream of the zinc-finger 438 protein binding site, which requires a zinc-finger protein for activation of transcription. Progesterone was dissolved in 2% dimethyl sulfoxide. Then, 10-µl reactions with different conditions were loaded into a 384-well plate. The mCherry fluorescence intensity was measured on a BioTek Synergy H1 Multimode Reader (Agilent) with an excitation wavelength of 565 nm and an emission wavelength of 615 nm at 34 °C for 8 h with 2-min intervals. All fits presented in Fig. 5 and Extended Data Fig. 4 were calculated from technical replicates (n = 3) using a nonlinear four-parameter curve-fitting analysis. All statistical analyses were based on two-way ANOVA with multiple comparisons.
Retrovirus production and primary murine T cell transduction
Retrovirus production and transduction of activated primary murine T cells were carried out as previously described76. Briefly, Phoenix-ECO cells (ATCC, CRL-3214) were seeded in a T125 flask and, after 48 h, transfected with polyethylenimine and plasmid mix. Cells were authenticated by the provider (STR genotyping) and tested negative for mycoplasma contamination (MycoAlert Mycoplasma Detection Kit, LT07-318). At 48 and 72 h after transfection, the supernatant containing the virus was collected, mixed, filtered through a 0.45-µm filter, concentrated using ultracentrifugation (24,000g, 2 h, 4 °C) and then stored at −80 °C.
Primary murine T cells were isolated from C57BL/6 mouse spleens using a specific isolation kit (Miltenyi Biotec, 130-095-130) and cultured in T cell medium (RPMI 1640 medium supplemented with GlutaMAX, 10 % (v/v) FBS, 100 U ml−1 penicillin, 100 µg ml−1 streptomycin sulfate, 1 mM sodium pyruvate, 50 µM 2-mercaptoethanol). Primary murine T cells were tested negative for mycoplasma (MycoAlert Mycoplasma Detection Kit, LT07-318). Cells were activated using αCD3/CD28 activation beads (11452D, Gibco) at a cell concentration of 0.5 × 106 cells ml−1 in T cell medium supplemented with 50 IU ml−1 of human IL-2 (200-02, PeproTech). Retroviruses were added to plates precoated with protamine and spun at 2,000g for 1.5 h at 32 °C. Activated T cells (0.5 × 106 cells per well) were transferred to each well. T cells were passaged 48 h posttransduction and maintained at 0.5 × 106 cells ml−1 in T cell medium supplemented with 10 ng ml−1 of human IL-7/IL-15 (200-7/200-15, PeproTech). Transduction efficiency was assessed by flow cytometry by measuring binding of a biotinylated HER2 protein (AcroBiosystems, HE2-H822R; 1:100 dilution) labelled with PE-conjugated streptavidin (Invitrogen, 12-4317-87; 1:100 dilution). For the transduction efficiency of the double chain, the chain containing FLAG-tagged Bcl2 and αHER2 was labelled with an A647-conjugated anti-FLAG antibody (Thermo Fisher, MA1−142-A647; 1:100 dilution), and the chain containing V5-tagged DBVen1619 was labelled with a fluorescein isothiocyanate (FITC)-conjugated anti-V5 antibody (GeneTex, GTX21209; 1:100 dilution).
Cytotoxicity assay of murine CAR-T cells
On day 10 after transduction, untransduced T cells, 2G-CAR-T cells and split CID-CAR-T cells (10 × 104) were cocultured with HER2-transduced MC38 mouse colon cancer cells (MC38-HER2; provided by L. Tang at EPFL) with an effector to target cell ratio of 1:1 in 96-well flat-bottomed plates. The number of CAR-positive cells was normalized to match the lowest transduction efficiency of the CID-CAR-T cells (Supplementary Fig. 16) by adding untransduced cells to achieve the same number of CAR-positive cells and maintain a consistent total cell count per well. Cytotoxicity activity of CAR-T cells was monitored for 48 h at different inducer concentrations. Target cells were labelled using Incucyte Nuclight Red to enable real-time counting of viable tumour cells with the IncuCyte live cell imaging system. All cell-based data presented in Fig. 5 were calculated from biological replicates (n = 3) and fitted using a nonlinear four-parameter curve-fitting analysis. MC38-HER2 cells were tested negative for mycoplasma (MycoAlert Mycoplasma Detection Kit, LT07-318).
Protein purification for crystallography
The 6xHis-tagged PDF1 from P. aeruginosa and DBAct553_1 were expressed in E. coli (BL21 T7 Express). Amino acid sequences of both proteins are shown in Supplementary Table 5. For PDF1, cells were grown in LB medium supplemented with 100 mM NiSO4 up to an OD600 of 0.7 at 37 °C, then induced with 1 mM IPTG and allowed to continue to grow overnight at 18 °C. For DBAct553_2, cells were grown in autoinduction medium up to an OD600 of 0.7 at 37 °C and then overnight at 18 °C. Cells were collected by centrifugation at 4,000g for 10 min, resuspended in lysis buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 5% glycerol, 1 mg ml−1 lysozyme, 1 mM PMSF and 1 µg ml−1 DNase) and lysed by sonication. Lysates were then clarified by centrifugation at 30,000g for 30 min and filtered. Proteins were purified using an ÄKTA Pure system (GE Healthcare) Ni-NTA HisTrap affinity column, followed by size-exclusion chromatography on a Superdex HiLoad 16/600, 75 pg, with Tris-buffered saline (50 mM Tris pH 7.5, 250 mM NaCl, 10 μM NiSO4) as a final buffer. PDF1, DBAct553_2 and actinonin were mixed at final concentrations of 35 μM, 105 μM and 300 μM, respectively, and incubated on ice for 1 h. Proteins were then concentrated by centrifugation before crystallization.
Crystallographic data collection and structure determination
The actinonin-bound PDF1–DBAct553_1 complex (5 mg ml−1) was crystallized using a sitting-drop vapour diffusion setup at 18 °C with 200 nl of protein and 200 nl crystallization solution consisting of 0.2 M sodium formate, 0.1 M sodium phosphate pH 6.2, 20% (v/v) PEG and 10% (v/v) glycerol. Crystals were cryoprotected with 25% glycerol and flash-cooled in liquid nitrogen. Diffraction data were collected at a temperature of 100 K at the European Synchrotron Radiation Facility (ESRF Grenoble, France). Raw data were processed and scaled with XDS (10 Jan. 2022, BUILT = 20220220) and then processed using the autoPROC package77 (GlobalPhasing, v.20230222). Phases were obtained by molecular replacement using the Phaser module of the Phenix package (v.1.20.1-4487) and a model from PDB 1LRY in complex with our designed binder DBAct553_1 (ref. 78). Atomic model adjustment and refinement were completed using COOT (v.0.9.5) and Phenix.refine79,80 (v.1.20.1-4487). Finally, MolProbity81 (v.4.5.1) was used to assess the quality of the refined model. Details of data collection and refinement statistics are shown in Extended Data Table 1.
Cryo-EM preparation and data acquisition
A chimeric DB3 Fab (Supplementary Table 5) was produced using the Expi293 expression system from Thermo Fisher Scientific (A14635). An anti-kappa light chain Fab82 (Supplementary Table 5) was produced using ExpiCHO-S cells (Thermo Fisher Scientific, A29127) growing in a ProCHO-5 medium (Lonza) supplemented with 2% dimethyl sulfoxide. Supernatants were collected 6 and 7 days, respectively, after transfection and filtered and purified by Ni-NTA affinity chromatography, followed by size-exclusion chromatography on a Superdex HiLoad 16/600, 75 pg. All proteins were concentrated in PBS as a final buffer. DBPro1156_2 was purified as described previously (‘Protein expression and purification’).
DB3 Fab, anti-kappa light chain Fab, DBPro1156_2 and progesterone were mixed with a molar ratio of 1:0.9:3:2, supplemented with 0.1% n-dodecyl-β-d-maltoside and concentrated to 3.87 mg ml−1. Proteins were applied to a glow-discharged 300-mesh holey carbon grid (Au 1.2/1.3, Quantifoil Micro Tools), blotted for 4 s at 95% humidity, 10 °C, plunge-frozen in liquid ethane (Vitrobot, Thermo Fisher Scientific) and stored in liquid nitrogen. Data collection was performed with automation program EPU (Thermo Fisher Scientific, v.2.12.1) on a 300 kV FEI Titan Krios G4 microscope equipped with a FEI Falcon IV detector. Micrographs were recorded at a calibrated magnification of ×120,000 with a pixel size of 0.658 Å and a nominal defocus ranging from −1.0 μm to −1.7 μm.
Cryo-EM image processing
Acquired cryo-EM data were processed (Supplementary Fig. 12) using cryoSPARC (v.4.4.1). Gain-corrected micrographs were imported, and micrographs with a resolution estimation worse than 5.5 Å were discarded after patch contrast transfer function estimation. A total of 16,038 micrographs were used for this complex. Initial particles were picked using a blob picker with 90–150-Å particle size. Particles were extracted with a box size of 360 × 360 pixels, downsampled to 140 × 140. After two-dimensional classification, clean particles were used for ab initio three-dimensional reconstruction. After several rounds of three-dimensional classification, the class with most detailed features was reextracted using full box size and subjected to non-uniform and local refinement to generate high-resolution reconstructions. The local resolution was calculated and visualized using ChimeraX83 (v.1.3, UCSF).
For structure building, we used ColabFold50 repredictions of the anti-kappa and DB3 Fabs, as well as the designed binder. Subsequent manual model adjustment and refinement were completed using Coot79 (v.0.9.5). Atomic model refinement was performed using Phenix.real_space_refine80 (v.1.20.1-4487). The quality of the refined model was assessed using MolProbity81 (v.4.5.1). Structural figures were generated using PyMOL (v.2.4, Schrödinger). The refined atomic models and corresponding cryo-EM maps were deposited under PDB accession code 9FKD and EMDB accession code EMD-50522. Details of data collection and refinement statistics are shown in Extended Data Table 2.
Chemical synthesis
All chemical reagents and solvents for synthesis were purchased from commercial suppliers (Sigma-Aldrich, Fluka, Acros) and were used without further purification or distillation. The composition of mixed solvents is given as a volume ratio (v/v). The 1H NMR spectra were recorded on a Bruker DPX 400 (400 MHz for 1H) with chemical shifts (δ) reported in ppm relative to the solvent residual signals (7.26 ppm for CDCl3; 3.31 ppm for MeOD) (Supplementary Fig. 17). Coupling constants are reported in Hz. Liquid chromatography coupled with mass spectrometry (LC–MS) was performed on a Shimadzu MS2020 connected to a Nexerra UHPLC system equipped with a Waters ACQUITY UPLC BEH Phenyl 1.7 µm 2.1 × 50 mm column. Buffer A consisted of 0.05% HCOOH in H2O; buffer B was 0.05% HCOOH in acetonitrile. The liquid chromatography gradient was as follows: 10% to 90% B within 6.0 min with 0.5 ml min−1 flow. Preparative high-performance liquid chromatography (HPLC) was performed on a Dionex system equipped with an UltiMate 3000 diode array detector for product visualization on a Waters SymmetryPrep C18 column (7 µm, 7.8 × 300 mm). Buffer A consisted of 0.1% v/v trifluoroacetic acid in H2O; buffer B was acetonitrile. The gradient was from 25% to 90% B within 30 min with 3 ml min−1 flow.
19-O-benzoyl-progesterone
First, 19-hydroxyprogesterone (2.0 mg, 6.1 µmol, 1 eq.) was dissolved in pyridine (0.5 ml); then, benzoyl chloride (0.9 µl, 7.9 µmol, 1.3 eq.) was added. The reaction mixture was stirred for 3 h. LC–MS analysis showed reaction completion, and 10 µl methanol was added. After 30 min, the solvents were evaporated under reduced pressure. The residue was dissolved in a minimum of acetonitrile and subjected to preparative HPLC. The fractions containing the product were pooled and lyophilized. The yield was 1.1 mg (41%). 1H NMR (400 MHz, CDCl3) δ 7.89 (d, J = 8.4 Hz, 2H), 7.56 (t, J = 7.4 Hz, 1H), 7.42 (t, J = 7.8 Hz, 2H), 5.98 (s, 1H), 4.81 (d, J = 11.3 Hz, 1H), 4.46 (d, J = 11.3 Hz, 1H), 2.68 (ddd, J = 17.0, 13.8, 5.9 Hz, 1H), 2.57–2.32 (m, 4H), 2.26–2.06 (m, 5H), 2.03–1.63 (m, 6H), 1.55–1.37 (m, 2H), 1.36–1.06 (m, 5H), 0.69 (s, 3H). HRMS (ESI/QTOF) m/z: [M + H]+ calcd for C28H35O4+ 435.2530; found 435.2528.
TBDMS-Act
Actinonin (2.0 mg, 5.2 µmol, 1 eq.) and 4-dimethylaminopyridine (3.8 mg, 31.2 µmol, 6 eq.) were suspended in dichloromethane (0.5 ml). TBDMS-Cl (2.5 mg, 16.6 μmol, 3.2 eq.) was added, and the reaction was stirred for 5 h at room temperature. The solvent was evaporated under reduced pressure, the residue was dissolved in MeOH (0.5 ml), water (50 µl) was added, and the reaction was heated to 60 °C for 5 h. The solvents were evaporated again, and the residue was dissolved in a minimum of dichloromethane and subjected to preparative thin-layer chromatography using dichloromethane/MeOH 9:1 as the eluent. The yield was 2.0 mg (77%). 1H NMR (400 MHz, MeOD) δ 4.38 (d, J = 8.5 Hz, 1H), 4.13 (s, 1H), 3.89 (dt, J = 10.0, 6.8 Hz, 1H), 3.79 (dd, J = 9.9, 5.3 Hz, 1H), 3.68 (dd, J = 9.9, 2.8 Hz, 1H), 3.63–3.42 (m, 1H), 2.83–2.75 (m, 1H), 2.34 (dd, J = 14.5, 8.0 Hz, 1H), 2.24–1.84 (m, 6H), 1.67–1.48 (m, 1H), 1.46–1.18 (m, 6H), 1.02–0.94 (m, 7H), 0.93–0.86 (m, 12H), 0.07 (s, 3H), 0.05 (s, 3H). HRMS (ESI/QTOF) m/z: [M+Na]+ calcd for C25H49N3NaO5Si+ 522.3334; found 522.3342.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.