Cloning, protein expression and purification
The complete open reading frames of SKI2 (SKIV2L, UniProt: Q6PGP7), SKI3 (TTC37, UniProt: Q15477) and SKI8 (WDR61, UniProt: Q9GZS3) were obtained from a human cDNA library (MegaMan Human Transcriptome Library, Agilent Technologies) and cloned in separate expression cassettes on a single plasmid (pACEBac1)35. SKI3 was designed with a cleavable 10ÃHis tag at the Nâterminus. The complex was expressed in baculovirus-infected Hi5 insect cells and purified by nickel affinity and ion-exchange chromatography. All derivatives, that is, SKI2N38, SKI2Îarch38 and SKI2Îwedge38, were expressed and purified similarly. A detailed protocol can be found in a previous study17.
The complete open reading frame of SKI7 (HBS1L isoform 3, UniProt: Q9Y450; also described as SKI7FL here) was commercially synthesized (Eurofins Genomics) and cloned with N-terminal 6ÃHis-TRX-3C and C-terminal GSG-eGFP-TwinStrep tags under an IPTG-inducible promotor for expression in Escherichia coli (pEC-A-6xHis-TrxA-3C-SKI7-eGFP-TwinStrepII construct). Transformed E. coli BL21 STAR pRARE cells were grown in TB medium at 37â°C under antibiotic selection to an optical density at 600ânm (OD600) of 2. The temperature was reduced to 18â°C and protein expression was induced by the addition of 500âμM IPTG. Cells were collected after 16âh by centrifugation at 8,500g and resuspended in lysis buffer containing 20âmM HEPES pH 7.5, 300âmM NaCl, 25âmM imidazole and 2âmM β-mercaptoethanol (β-ME), and supplemented with 200âUâmlâ1 benzonase (Merck), 500âμM AEBSF protease inhibitor and cOmplete EDTA-free protease inhibitor cocktail (Roche). The cells were lysed by ultrasonication (Bandelin, Sonopuls basic). The recombinant protein was kept strictly at 4â°C and the purification procedure was performed quickly to avoid degradation. The lysate was cleared by centrifugation and loaded on a HisTrap HP 5-ml column (Cytiva) for nickel affinity chromatography. The column was washed with 15 column volumes of 20âmM HEPES pH 7.5, 1,000âmM NaCl, 200âmM KCl, 10âmM MgCl2, 25âmM imidazole and 2âmM β-ME, followed by washing with 5 column volumes of lysis buffer. The protein was eluted with lysis buffer supplemented with 350âmM imidazole into a StrepTrap HP 5-ml column (Cytiva) for a second affinity step. The bound protein was washed with 10 column volumes of buffer containing 20âmM HEPES pH 7.5, 300âmM NaCl and 2âmM DTT, and the addition of 5âmM desthiobiotin (DTB) eluted the protein. The protein was concentrated in 10% glycerol (v/v), flash frozen in liquid nitrogen (LN2) and stored at â80â°C.
The SKI7SKI-EXO construct (residues 369â632) was cloned with a N-terminal 6ÃHis-TRX-3C tag and also with a C-terminal 3C-eGFP-TwinStrep tag. The construct behaved similarly to SKI7FL and was quickly purified with the same protocol to prevent degradation.
The shorter SKI7 constructs (residues 1â144 (here also SKI740S), 136â277, 267â386, 372â546 (here also SKI7SKI) and 540â632 (here also SKI7EXO)) were cloned similarly with N-terminal 6ÃHis-TRX-3C and C-terminal GSG-eGFP-TwinStrep tags. The constructs expressed well and had no tendency for degradation. An initial affinity chromatography step followed by cleavage of the N-terminal 6ÃHis-TRX tag during overnight dialysis combined with a second nickel affinity step to remove the cleaved 6ÃHis-TRX tag resulted in sufficiently pure constructs as judged by Coomassie-stained SDSâPAGE. The N-terminal 6ÃHis-TRX tag of SKI7EXO (residues 540â632) was not cleaved, because its presence helped to avoid protein precipitation during the final concentration step. For cryo-EM sample reconstitution, TRXâSKI7SKI was also purified without cleavage of the TRX tag.
The complete open reading frame of DIS3L (UniProt: Q8T64F) was cloned with a cleavable TwinStrep tag at the Nâterminus into the piggyBac vector and expressed in a HEK 293T stable cell line adapted to grow in suspension. In brief, 400â800âml of stably transfected cells (106âcells per ml) were induced with 1âμgâmlâ1 doxycycline for 48âh and collected by centrifugation at 800g. Cells were resuspended in lysis buffer containing 20âmM HEPES pH 7.5, 300âmM NaCl and 2âmM DTT, and lysed using a dounce homogenizer. After clearing the lysate by centrifugation, it was loaded on a 5-ml StrepTrap XT column (Cytiva) and washed with high salt (20âmM HEPES pH 7.5, 1,000âmM NaCl, 200âmM KCl, 10âmM MgCl2 and 2âmM DTT) and lysis buffer. The protein was eluted in buffer containing 20âmM Tris-HCl pH 8.0, 150âmM NaCl, 50âmM biotin and 2âmM DTT. A detailed description, including generation of the HEK 293T stable cell line, can be found in a previous study17.
Reconstitution of cytoplasmic exosome
Human cytoplasmic exosome (EXO10) was reconstituted from equimolar amounts of pre-assembled EXO9 (purified as previously described22,28 and freshly purified DIS3L or DIS3L(D486N). After incubation for 30âmin on ice, the mixture was concentrated by centrifugation at 3,000g using an Amicon Ultra MWCO100 centricon (Millipore) and purified by size-exclusion chromatography (Superdex 200 Increase 10/300 GL, Cytiva) in buffer containing 20âmM HEPES pH 7.5, 150âmM NaCl and 2âmM DTT.
Purification of ribosomal subunits
Human ribosomal 40S and 60S subunits were obtained from HEK 293T cells (using adapted protocols36,37). In brief, ribosomes were pelleted by ultracentrifugation from cleared HEK 293T cell lysate and resuspended in buffer containing 2âmM puromycin and 500âmM KCl to release the nascent peptide chain and dissociate the ribosomal subunits. The 40S and 60S ribosomal subunits were subsequently separated by sucrose gradient centrifugation and the corresponding 40S and 60S fractions concentrated in buffer containing 20âmM HEPES pH 7.5, 50âmM KCl, 4âmM MgCl2 and 2âmM DTT. A detailed description can be found in a previous study17.
Co-precipitation assays
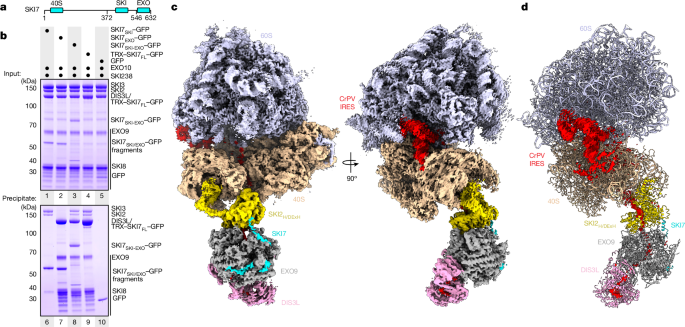
A total of 100âpmol GFP-tagged SKI7FL or derivates (SKI740S 1â144, SKI7 136â277, SKI7 267â386, SKI7SKI 372â546 and SKI7EXO 540â632) were mixed with 130âpmol SKI238, SKI2N38 or EXO10 in buffer containing 20âmM HEPES pH 7.5, 150âmM NaCl, 0.05% v/v NP40 and 2âmM DTT. After incubation with GFP-binder resin (ChromoTek GFP-Trap Agarose) at 4â°C for 45âmin and three washes in the same buffer, the resin was dried and taken up in SDS buffer. Input and precipitate fractions were analysed by Coomassie-stained SDSâPAGE on a NuPAGE 4â12% Bis-Tris gel (Thermo Fisher Scientific). Co-precipitation of 40S and 60S ribosomal subunits was done accordingly, but in buffer containing 20âmM HEPES pH 7.5, 50âmM KCl, 4âmM MgCl2, 0.05% v/v NP40 and 2âmM DTT.
Intensity-based absolute quantification analysis of proteomic data
Intensity-based absolute quantification (iBAQ) values were collected across five cell lines (GM12878, hESC, HeLaS3, HepG2 and K562) using five different proteases (AspN, GluC, LysC, LysN and trypsin) from a previous proteome study26. The data were plotted as box plots in Extended Data Fig. 1b. Here, quartiles are represented by boxes with whiskers extending to the highest or lowest value within 1.5 times the interquartile range. Median values are shown as thick horizontal bars inside the boxes. Values falling further than 1.5âtimes the interquartile distance are considered outliers and are shown as dots. iBAQ values are plotted in log scale.
In vitro transcription of RNA substrates
The genomic sequence of the CrPV-IRES was taken from the intergenic region of the CrPV genome (NCBI GeneBank: 6025â6232ânt, NC_003924.1) and fused to a portion of the firefly luciferase open reading frame (modified from a previous report38). The used CrPV-IRES construct was amplified by PCR, gel-purified and transcribed in vitro with T7 RNA polymerase by run-off transcription. The transcription reaction was performed in 40âmM Tris-HCl pH 8.0, 28âmM MgCl2, 0.01% Triton X-100, 1âmM spermidine and 5âmM DTT in the presence of 25âmM of each ribonucleotide (ATP, CTP, GTP and UTP; Jena Bioscience) and 100âUâμlâ1 T7 RNA Polymerase (MPI Biochemistry Core Facility) at 37â°C for 4âh. The DNA template was digested with 1âUâμlâ1 DNase I (Roche) and the CrPV-IRES purified by LiCl2 precipitation. A detailed description can be found in a previous study17.
The transcription template for the Brc-CrPV-IRES used in the exosome degradation assays was amplified by PCR, fusing the sequence of the broccoli light-up aptamer (GCGGAGACGGTCGGGTCCAGATATTCGTATCTGTCGAGTAGAGTGTGGGCTCCGC)39 to the 5â² end of the CrPV-IRES. Otherwise, the Brc-CrPV-IRES was transcribed as described above.
Exosome RNA-degradation assays
The 80S-Brc-CrPV-IRES substrate used in the assays was reconstituted by mixing 100âpmol purified human 40S with 120âpmol Brc-CrPV-IRES. After incubation at 37â°C for 5âmin, 120âpmol purified human 60S was added, and the incubation was allowed to proceed for another 10âmin. The 80S-Brc-CrPV-IRES complex was subsequently purified by sucrose gradient centrifugation in a SW 40 Ti rotor (Beckman Coulter) at 4â°C. Sucrose gradients (15â40%) (w/v) in buffer containing 20âmM HEPES pH 7.5, 50âmM KCl, 4âmM MgCl2 and 2âmM DTT were mixed using the Gradient Master (Biocomp). After 17âh centrifugation at 22,800ârpm, the gradients were fractionated using a Piston Gradient Fractionator (Biocomp). Fractions corresponding to the 80S-Brc-CrPV-IRES complex were concentrated in buffer containing 20âmM HEPES pH 7.5, 50âmM KCl, 4âmM MgCl2 and 2âmM DTT by centrifugation at 3,000g using an Amicon Ultra MWCO100 centricon (Millipore). The concentrated 80S-Brc-CrPV-IRES was estimated to be 0.24âμM and used immediately.
Exosome degradation assays were performed in technical triplicates, over a time course of 45âmin at 37â°C in buffer containing 20âmM HEPES pH 7.5, 50âmM KCl, 4âmM MgCl2 and 2âmM DTT, supplemented with 0.5âmM ADP or ATP as indicated in the condition. A single reaction contained 2âpmol 80S-Brc-CrPV-IRES substrate, 2âpmol SKI238, 4âpmol SKI7 and 4âpmol EXO10 in a total volume of 30âμl. Reactions without the addition of EXO10 were included as negative controls for every condition. Time-point samples were taken at 0âmin, 5âmin, 15âmin and 45âmin, and the reaction was stopped by adding 10à excess buffer containing 100âmM Tris-HCl pH 7.5, 150âmM NaCl, 300âmM NaOAc pH.5.2, 10âmM EDTA and 1% SDS. The reactions were purified by phenol extraction and ethanol precipitation and separated on a denaturing 12% polyacrylamide gel containing 7âM urea. After incubation of the gel in water to remove urea, the Brc-CrPV-IRES was refolded for 20âmin under agitation in the presence of 3,5-difluoro-4-hydroxybenzylidene imidazolinone (Sigma-Aldrich) in buffer containing 40âmM HEPES pH 7.5, 125âmM KCl and 10âmM MgCl2, and subsequently visualized by fluorescence at a 501-nm wavelength (Typhoon FLA7000, Cytiva). Gel images were quantified by densitometry in ImageJ and plotted with R.
Cryo-EM sample preparation
The exosomeâribosome supercomplex sample (EXO10-SKI-80S) was prepared by sequentially mixing the individual components, similar to the ribosome-bound samples that we previously described17. Sucrose gradient centrifugation and concentration steps resulted in reduced amounts of 80S-bound exosomes according to SDSâPAGE and cryo-EM analysis, and were therefore omitted from the sample preparation procedure. In brief, 100âpmol of purified human 40S was mixed with 100âpmol CrPV-IRES carrying a 29-nt 3â² overhang (referred to throughout the text simply as CrPV-IRES) in buffer containing 20âmM HEPES pH 7.5, 50âmM KCl, 4âmM MgCl2 and 2âmM DTT. After incubation at 37â°C for 5âmin, 110âpmol human 60S was added and incubation proceeded for another 10âmin. Next, 100âpmol of purified wild-type SKI238 was added to the 80S-CrPV-IRES complex and incubated for 5âmin at 37â°C before adjusting the reaction to 1âmM ATP. After incubation at 37â°C for 15âmin, 100âpmol of preincubated SKI7SKI-EXOâEXO10(D486N) (equimolar amounts of EXO10(D486N) and SKI7SKI-EXO incubated for 15âmin at 37â°C) was added to the reaction. After 5âmin at 37â°C the reaction was adjusted to 2âmM ADP-BeF to stop the ATPase reaction. The incubation was allowed to proceed for another 15âmin before adjusting the final volume to 130âμl with the buffer described above and storing the sample at 4â°C. The concentrations of SKI238, SKI7SKI-EXO and EXO10(D486N) were between 12 and 20âμM, leading to only minor alterations in volume and salt concentration in the reaction, and were not compensated for. Aggregated protein was removed by centrifugation at 10,000g and the sample supplemented with 0.04% (v/v) n-octyl-β-d-glucoside before grid preparation. Two times 4-5âμl of the sample were incubated for 1âmin on holey carbon grids with carbon support (R2/1, 200 mesh, Quantifoil), that were previously glow-discharged with negative polarity at 20âmA for 30âs using an EMS GloQube (MiTeGen). The sample was plunge-frozen in a liquid ethaneâpropane mix using a Vitrobot Mark IV (Thermo Fisher Scientific) operated at 4â°C and 95% humidity.
The SKI7-SKI238 sample was prepared by mixing 600 pmol SKI2Îarch38 with approximately 900âpmol of TRXâSKI7SKI and incubating at 37â°C for 20âmin in the presence of 3C protease to cleave the TRX tag. The SKI7SKI fragment (residues 372â546) was difficult to quantify because it does not contain amino acids that would absorb at 280ânm. Therefore, the TRX tag was kept and only cleaved while reconstituting the complex. The sample was then concentrated in buffer containing 20âmM HEPES pH 7.5, 100âmM NaCl and 2âmM DTT to approximately 30âμl. After centrifugation at 18,000g for 10âmin to pellet larger aggregates, the sample was injected onto a Superose 6 Increase 3.2/300 column (Cytiva) for size-exclusion chromatography by an Aekta micro (Cytiva). A single peak fraction, containing approximately 500ânM, was collected, and mixed with 0.04% (v/v) n-octyl-β-d-glucoside. Four to five microlitres of the sample was applied to holey carbon grids (R2/1, 200 mesh, Quantifoil), that had been glow-discharged with negative polarity at 20âmA for 30âs using an EMS GloQube (MiTeGen) beforehand. The sample was plunge-frozen in a liquid ethaneâpropane mix using a Vitrobot Mark IV (Thermo Fisher Scientific) operated at 4â°C and 95% humidity.
Cryo-EM data collection and processing
The cryo-EM data of the exosomeâribosome supercomplex were collected on a FEI Titan Krios GIII microscope (Thermo Fisher Scientific) at 300âkV, equipped with a Gatan K3 direct electron detector operating in electron-counting mode. The microscope is equipped with a post column energy filter (Gatan GIF Quantum LS) set to a slit width of 10âeV. Images were collected by underfocused acquisition (target range of â0.6âμm and â2.4âμm) at a nominal magnification of 105,000à set up in SerialEM40 using a beam-tilt-based multi-shot acquisition scheme for faster imaging. This resulted in 48,004 micrograph movies (30 movie frames each) acquired at a pixel size of 0.85âà per pixel with a total exposure of 64.14âe per à 2 spread over 3âs.
On-the-fly micrograph movie processing was assisted by Focus41, which ran MotionCor2 (ref. 42), GCTF43 and Gautomatch (https://www2.mrc-lmb.cam.ac.uk/download/gautomatch-053/) on individual images while the data were being collected. Subsequent particle processing was performed in RELION v.3.1 (ref. 44). Eightfold downscaled particles were extracted from the aligned, exposure-weighted micrographs and classified in 2D and 3D to discard non-ribosomal and low-resolution particles from the data. These data were separated on the basis of the results of the 2D classification into one particle stack displaying features of the EXO10-SKI-80S assembly and a second one showing density for the EXO10-SKI-40S assembly only.
The cleaned EXO10-SKI-80S particles (1,189,927) were extracted at the original pixel size of 0.85âà per pixel and aligned in 3D auto-refinement with a spherical mask using a 40-à downfiltered starting model based on PDB 4UG0 (ref. 45). The refined particles were then used to subtract a large portion of the 80S ribosome signal from the corresponding particle images. To improve the alignment precision and the quality of the reconstructions in the subsequent steps, the particle images were recentred on the remaining EXO10-SKI signal and re-extracted in a smaller box in RELION v.3.1 (ref. 44). Subtracted particle images that did not show SKI, EXO10 or leftover ribosome signal were removed in 3D classification and resulted in a set of 565,489 cleaner particles for downstream processing. Further classification in 3D showed a small range of different particles which aligned in classes with SKI2H complexed by a prominent EXO10 density, with SKI238 without visible exosome density, and in classes that only showed leftover ribosome signal without visible EXO10-SKI density.
In a final step, all EXO10-SKI-containing particles (229,580) were 3D-classified into 6 classes, which resulted in 79,353 well-aligning particles. The ribosome-signal-subtracted particles were aligned by 3D auto-refinement and resulted in a reconstruction of EXO10-SKI at an overall resolution of 3.7âà (according to the Fourier shell correlation (FSC) cut-off criterion of 0.143)46. This map is referred to as the âEXO10-SKI-80S (80S subtracted)â map, or map1. For masking and map sharpening in the RELION v.3.1 post-processing procedure44, an ad hoc b-factor of â30 was applied. The automatically determined b-factor of â110.8 resulted in an oversharpened map with a loss of connectivity particularly in the area of DIS3L. Next, the signal subtraction was reverted and the corresponding EXO10-SKI-80S particles refined to 3.2-à global resolution (according to the FSC cut-off criterion of 0.143)46. An ad hoc b-factor of â20 was preferred over the automatically estimated b-factor of â71.7 in the RELION post-processing routine44 to better visualize the 40S ribosomal subunit. This reconstruction is referred to as the âEXO10-SKI-80S (full map)â map, or map3. The quality of the map in areas of the CrPV-IRES and EXO10-SKI, however was not satisfactory. Therefore, we subtracted the signal of the 60S ribosomal subunit from the reconstruction and aligned the resulting EXO10-SKI-40S particles by 3D auto-refinement, which yielded a reconstruction at 3.3-à global resolution (according to the FSC cut-off criterion of 0.143)46. Masking and map sharpening in RELION post-processing44 was performed using an ad hoc b-factor of â20 (automatically determined b-factor â73.6). Although the map quality for EXO10-SKI in this reconstruction improved only marginally, the resolution and volume connectivity for the CrPV-IRES in the intersubunit space improved substantially. This EXO10-SKI-40S reconstruction is referred to as the âEXO10-SKI-80S (60S subtracted)â map, or map2.
For display and visualization purposes, a composite map of the EXO10-SKI-80S assembly was calculated using map1, map2 and map3 and is shown in Fig. 1. The EXO10-SKI map detailed in Fig. 2 and Extended Data Fig. 3 was produced using DeepEMhancer47 with the wide target setting.
As described above, the ribosome-bound EXO10-SKI supercomplex data contained a fraction of particles that lacked the entire 60S ribosomal subunit (described as EXO10-SKI-40S here). Two- and three-dimensional classification was performed in RELION v.3.1 (ref. 44) and resulted in 119,144 clean 40S ribosomal particles, which were aligned in 3D auto-refinement for subsequent subtraction of the majority of the 40S ribosome signal. The subtracted particle images were 3D-classified into four classes using a wide mask and a 30-à low-pass-filtered starting model of the SKI2N38 gatekeeping module (PDB 7QDS)17. The classification showed one class with 53,460 particles that shows well-aligning density for the SKI2N38 gatekeeping module. Three-dimensional auto-refinement of the subset gave a reconstruction at 4.1-à global resolution (according to the FSC cut-off criterion of 0.143)46. An ad hoc b-factor of â30 was used in the RELION v.3.1 post-processing procedure (automatically determined b-factor â134.4)44. This reconstruction is referred to as âEXO10-SKI-40S (EXO10-SKI2Hâ+â40S subtracted)â, or map5. For the corresponding ribosome-bound reconstruction, signal reversion procedures similar to the EXO10-SKI-80S particles above were applied, which led to a EXO10-SKI-40S reconstruction at 3.4-à global resolution using a manually determined b-factor of â20 (automatically determined b-factor â68.9). This reconstruction is labelled âEXO10-SKI-40S (full map)â, or map6. Although the reconstruction shows clear density for the SKI2N38 and SKI2H helicase modules interacting at two different positions of the 40S subunit, the signal for EXO10 was less coherent. To improve the EXO10 density, repeated signal subtraction of the 40S ribosomal subunit combined with 3D classification on the exosome and signal reversion resulted in 40S ribosome signal-subtracted and signal-reverted 3D reconstructions at 6.5âà (ad hoc b-factor â30, automatically determined b-factor â128.3) and 4.7âà (ad hoc b-factor â20, automatically determined b-factor â92.5) global resolution, respectively. The reconstruction focused on EXO10-SKI is labelled âEXO10-SKI-40S (40Sâ+âSKI2N38 gatekeeping module subtracted)â, or map7, and the signal-reverted reconstruction is named âControl EXO10-SKI-40Sâ, or map8. The main purpose of the latter was to clarify that in the particle stack showing clear presence for the EXO10-SKI assembly, the SKI2N38 gatekeeping module is also bound to the 40S subunit (see also Extended Data Fig. 7g).
For display and visualization purposes, a composite map of the EXO10-SKI-40S assembly was calculated using map5, map6 and map7 and is shown in Fig. 3.
Data processing of the ribosome-bound human EXO10-SKI supercomplex dataset required signal subtraction of the ribosome to yield reconstructions of comparable interpretability and quality to those described in the manuscript. Classical focused classification and refinement procedures did not yield results of comparable quality.
The EXO10-SKI-80S particles show additional density at the location where the SKI2N38 gatekeeping module binds to the 40S ribosomal subunits in the particles that lack 60S (see Extended Data Fig. 6b). However, the signal is rather weak in comparison, owing presumably to the presence of the 60S. To improve the additional unaccounted density at the 40S head (Extended Data Fig. 6b, left) focused 3D classification using a spherical mask located around the area of interest was performed. The resulting particle stack was used for 3D refinement, resulting in a 3D reconstruction of the 80S ribosome. A low-pass-filtered map of this assembly was used to place the model of SKI2N38 by rigid-body fitting (Extended Data Fig. 6b, right).
The cryo-EM data of the human SKI7-SKI238 complex were collected similarly to the ribosome-bound dataset described above. A total of 20,224 micrograph movies were acquired at a pixel size of 0.85âà per pixel with a total exposure of 67.8âe per à 2 spread over 6âs and 40 movie frames. Beam-induced motion correction, CTF estimation, particle picking and processing were done similarly to what was done for the ribosome-bound data above. Fourfold downscaled particles were first extracted from the aligned, exposure-weighted micrographs, and particles that seemed to be non-SKI238 were discarded by 2D and 3D classification. The resulting subset of 422,913 SKI238 particles was extracted at the original pixel size of 0.85âà per pixel. Further 3D classification into six classes, using a starting model based on a previous cryo-EM structure of the SKI238 complex (PDB 7QDS)17, gave a final subset of 151,860 particles. Initial 3D auto-refinement gave a 3D reconstruction at an estimated overall resolution of 3.6âà . The quality of the reconstruction and the level of resolved detail were further improved by Bayesian polishing to correct for per-particle motion and by iterative refinement of the per-particle CTF (taking into account the beam-tilted data-acquisition scheme). This resulted in a final reconstruction with an overall global resolution of 3.4âà according to the gold-standard FSC (0.143) criterion46. Masking and map sharpening using an automatically determined global b-factor of â99.4 were performed in the RELION post-processing routine44. This map is referred to here as âSKI7-SKI238â, or map4.
Density interpretation, model building and refinement
The 80S-bound EXO10-SKI or map1 reconstruction was interpreted by rigid-body fitting pre-existing models of SKI2 (PDB 7QE0)17, the nuclear human exosome (PDB 6D6Q)27 and AlphaFold multimer predictions of either SKI7EXO or DIS3L with different human exosome subcomplexes48. Initial rigid-body fitting was performed in UCSF Chimera49. In several areas of the map (the EXO9 barrel and SKI2H), the resolution and quality of the reconstruction allowed us to manually adjust the fit of the models in Coot50, followed by real-space refinement from within the PHENIX suite51. In peripheral areas of the reconstruction (the DIS3L exonuclease), the resolution and quality of the map was not as good, and required us to use a focused refined map with improved map quality and connectivity, which allowed for rigid-body docking of the AlphaFold model. The electron density for the RNA path inside SKI2H, between SKI2H and EXO9, and within DIS3L was sufficient to model individual nucleotides. However, the precise sequence of the RNA inside the barrel could not be determined from the reconstruction. Inside the EXO9 barrel and between EXO9 and DIS3L, a previous model (PDB 6D6Q)27 was used to inform structure building. The final model of the 80S-bound EXO10-SKI complex was validated using MolProbity52.
The complete model of 80S-bound EXO10-SKI was subsequently assembled by rigid-body fitting of the individual 80S, CrPV-IRES RNA and the above-described EXO10-SKI model into the full reconstruction of the EXO10-SKI-80S (map3) in PHENIX. As a starting model for the human 80S-CrPV-IRES, we used PDB 4UG0 (ref. 45) and PDB 4V92 (ref. 36) as already described in detail previously17. After placing the models into the map, the CrPV-IRES was manually adjusted and refined in PHENIX to reflect the lack of PK-1, the connection between the RNA exit of the ribosome and entry into SKI2H and the channelling through the EXO10-SKI complex.
The model for the reconstruction of the 40S-bound SKI2N38 gatekeeping module from which the signal of the 40S subunit and EXO10-SKI had been subtracted (map5) was interpreted with a pre-existing model of the SKI2N38 gatekeeping module (PDB 7QDS)17. Initial rigid-body fitting was performed in UCSF Chimera49. Inspection of the fit in Coot50 showed insufficient density for several TPRs towards the Nâterminus of SKI3, which were consequently not modelled. Otherwise, the model is in good agreement with the experimental density. The final model was real-space-refined within the PHENIX suite51 and subsequently validated using MolProbity52.
The model for the reconstruction of the 40S-bound EXO10-SKI from which the signal of the 40S and the SKI2N38 gatekeeping module had been subtracted (map7) was interpreted by rigid-body fitting of the above-described 80S-bound EXO10-SKI model, real-space-refined within the PHENIX suite51 and validated using MolProbity52.
The complete model of the 40S-bound EXO10-SKI was subsequently assembled by rigid-body fitting of the individual models of the 40SÂ subunit, the adjusted model of the CrPV-IRES similar to that described above, the model of the SKI2N38 gatekeeping module and the EXO10-SKI model into the full reconstruction of the 40S-EXO10-SKI (map6) in PHENIX. As a starting model for the human 40S, we used the model of the 40S ribosomal subunit from PDB 4UG0 (ref. 45).
The reconstruction of SKI7-SKI238 (map4) was interpreted with pre-existing models of the SKI2N38 gatekeeping module (PDB 7QDS)17 and an AlphaFold multimer prediction of a part of the SKI2N38 gatekeeping module in complex with SKI7SKI. Initial rigid-body fitting was done in UCSF Chimera49. The resolution and quality of the map allowed us to manually modify and refine the models in Coot50 and subject them to subsequent rounds of real-space refinement from within the PHENIX suite51. The final model of human SKI7-SKI238 was validated using MolProbity52.
Isolation of stalled RNCs and polysome profiling
The 5â² capped XBP1-XTEN mRNA was obtained by an in vitro transcription reaction from the corresponding DNA template using the mMESSAGE mMACHINE T7 transcription Kit (Thermo Fisher Scientific) as described previously33. In vitro translation reactions were performed using the rabbit reticulocyte lysate (RRL) system (nuclease treated, Promega). Translation of the 5â² capped XBP1-XTEN mRNA was performed at 32â°C for 25âmin using 0.05âμM XBP1-XTEN template and 0.8âUâμlâ1 RNasin Plus ribonuclease inhibitor (Promega) in a total reaction volume of 1âml. The stalled RNCs on the 5â² capped XBP1-XTEN mRNA were affinity-purified through the 8ÃHis tag on the nascent polypeptide chain and magnetic beads, as described33. The amounts of isolated ribosomes were estimated by UV absorbance at 260ânm. Sucrose density gradients (10â50% sucrose in 50âmM HEPES pH 7.5, 100âmM KOAc, 5âmM Mg(OAc)2, 2âmM DTT, 0.1% octaethylene glycol monodedecyl ether (TCI) and 20âUâmlâ1 recombinant RNasin ribonuclease inhibitor (Promega)) were prepared using a Gradient Master (Biocomp). The purified RNCs were incubated with 1.2 à molar excess of SKI2N38 or SKI2Îwedge38 for 40âmin at 4â°C, subsequently applied on a sucrose gradient, and the ribosomal fractions were separated by centrifugation for 3âh at 172,000g and 4â°C in an SW 40 Ti rotor (Beckman Coulter). Polysome profiles were generated by continuous absorbance measurement using an UV absorbance reader (Gilson) and simultaneously fractionated (around 3.39âmm per fraction) with a Piston Gradient Fractionator (Biocomp). For display purposes, the sucrose gradient profiles were superimposed using the highest A260 maximum of the disome peak as a reference.
Western blotting
Fractions obtained by gradient fractionation were precipitated with trichloroacetic acid and pellets were resuspended in SDS loading buffer. All samples were analysed on 15% SDSâPAGE followed by western blotting onto Immobilon-PSQ transfer membrane (Merck Millipore). Transfer efficiency was monitored by stain-free imaging using the 2,2,2-trichloroethanol in-gel technique. Western blots were probed against monoclonal anti-Flag antibody (Sigma-Aldrich, F3165) in a dilution of 1:5,000 and polyclonal anti-mouse HRP-coupled antibody (Bio-Rad, 172-1011) in a dilution of 1:10,000 in 5% milk, made from 5% w/v milk powder (Roth) in 1Ã Dulbeccoâs PBS buffer (DPBS, Thermo Fisher Scientific) and 0.001% Tween20 (Bio-Rad). The chemiluminescence signal was detected using Amersham ECL prime western blotting detection reagent (Cytiva) on a ImageQuant LAS 4000 (GE Healthcare; exposed for 140âs at standard sensitivity).
Statistics and reproducibility
All in vitro assays (protein co-precipitation (Fig. 1b and Extended Data Figs. 1c,d and 4h); protein purifications and complex reconstitutions (Extended Data Figs. 2a and 4a)), in part or in whole, were successfully reproduced at least twice. The nuclease assays (Extended Data Fig. 1f) were performed as triplicates. For each cryo-EM sample (Extended Data Figs. 2b and 4b), multiple grids were screened before final data collection. The final datasets presented in this article were collected once. The sucrose gradient centrifugations and corresponding western blots (Fig. 4b and Extended Data Fig. 9b) were reproduced twice.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.