Cloning
Cloning of S. pombe Uba1, Cys-Ub with an N-terminal Ulp1-cleavable His6âSmt3 tag, and Ub without tags was described previously15,17,52. All mutations were generated using PCR-based mutagenesis. Constructs encoding full-length S. pombe Ubc4(C21S/C107S) with a C-terminal GlyâGly linker followed by thrombin-cleavable His6 tag, and constructs for full-length S. pombe Ubc4 with C-terminal His6 tag were constructed using the pET29b vector (Novagen). The DNA encoding S. pombe Pub2HECT (amino acid residues 295â671) was inserted into the pSMT3 vector to introduce an N-terminal Ulp1-cleavable53 His6 Smt3 tag. The DNA encoding S. pombe Ub lacking the last two amino acids at its C terminus, with StrepTag and TEV protease cleavage site (MWSHPQFEKSAENLYFQGSGG) added at its N-terminus, further referred to as Ub(â2), was inserted into vector pTXB1 (New England Biolabs), to generate a C-terminal fusion with an Mxe intein-chitin binding domain.
Protein expression and purification
DNA plasmids encoding recombinant proteins were expressed in Escherichia coli strain BL21 (DE3) codon plus (Stratagene). To produce Pub2HECT, cells were grown in Superbroth at 37â°C to OD600â=â0.8, cooled in an ice/water bath for 20âmin before addition of 1-thio-β-d-galactopyranoside (IPTG) to a final concentration of 0.4âmM. Cultures were incubated for 14â18âh at 18â°C. Uba1, Ubc4, Cys-Ub and untagged Ub were expressed as described previously15,17,52. Cells were collected by centrifugation at 4,000g (Beckman JLA-8.1000) for 20âmin at 4â°C. Cell pellets were resuspended in 50âmM sodium HEPES pH 8.0, 350âmM NaCl, 20% sucrose and snap frozen in liquid nitrogen. Cell pellets were thawed, supplemented in lysis buffer containing 0.5âmM TCEP, 2.5âmM MgCl2, 0.1âmgâmlâ1 DNAse I, 1âmgâmlâ1 lysozyme, 1âmM PMSF, and lysed by sonication (2âs on, 8âs off, 50% output amplitude) twice for 3âmin with a 3âmin break in between using Digital Sonifier 450 Cell Disruptor (Branson) on ice/water bath. Lysates were clarified by centrifugation at 47,000g (Beckman JA-20).
For purification of His6-tagged proteins, supernatant lysates were mixed with 15âmM imidazole and applied to Ni2+-NTA superflow resin (Qiagen) by gravity flow. Beads were washed with buffer containing 20âmM sodium HEPES pH 8.0, 350âmM NaCl, 0.2âmM TCEP, and 20âmM imidazole at 4â°C. Proteins were eluted in buffer containing 20âmM sodium HEPES pH 8.0, 350âmM NaCl, 0.2âmM TCEP, and 250âmM imidazole at 4â°C. His6âSmt3 tags used for affinity purification were removed by incubation with Ulp153 and separated by size-exclusion chromatography (SEC) with Pub2 and Ubc4 separated using HiLoad 26/600 Superdex 75 PG column and Uba1 separated using HiLoad 26/600 Superdex 200 PG column (GE Healthcare), equilibrated in 20âmM sodium HEPES pH 7.5, 250âmM NaCl, 0.1âmM TCEP at 4â°C. For Ubc4(C21S/C107S), the C-terminal His6 tag was removed by treatment with thrombin then separated by SEC. After SEC, Uba1 was further purified by anion-exchange chromatography using a MonoQ 10/100 GL column (GE Healthcare) in 20âmM sodium HEPES pH 8.0, 0.1âmM TCEP and a linear gradient of 50â800âmM NaCl at 4â°C. To obtain fluorescein-Ub variants, Cys-Ub variants containing an additional N-terminal Cys for labelling were obtained as described previously15. Purified Cys-Ub variants were incubated in 20âmM HEPES pH 7.5, 200âmM NaCl, 2âmM TCEP for 10âmin at room temperature, desalted into the same buffer without TCEP and modified by adding 10-fold molar excess of 5-fluorescein maleimide (Thermo Fisher Scientific) and incubating for 2âh at room temperature. Reactions were then buffer exchanged to 20âmM sodium HEPES pH 7.5, 200âmM NaCl using 7âkDa MWCO Zeba Spin Column (Thermo Fisher Scientific) and separated by size exclusion using Superdex 75 Increase 10/300 GL column (GE Healthcare) equilibrated in the same buffer at 4â°C. Native Ub (without affinity tags), was purified as described previously52, with additional SEC using a HiLoad 26/600 Superdex 75 prep grade column (GE Healthcare) equilibrated in 20âmM Trisâ¢HCl pH 7.5, 250âmM NaCl at 4â°C. Fractions containing the desired proteins were pooled, concentrated, flash-frozen in liquid nitrogen, and stored at â80â°C.
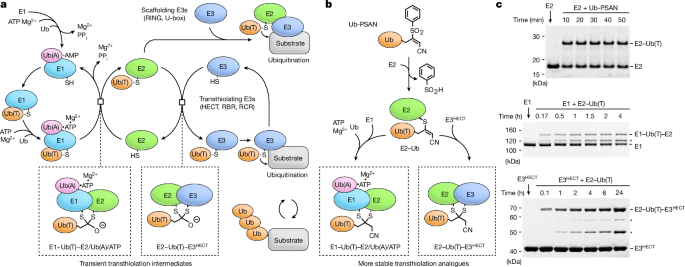
Preparation of Ub-PSAN
H2N-GlyâPSAN was synthesized as described28. Ub1â74 fused to Mxe intein-chitin binding domain was expressed, and cells collected and lysed using the same procedure as for Pub2HECT. Lysate supernatant was incubated with chitin beads (New England BioLabs) equilibrated in 30âmM sodium HEPES pH 8.0, 350âmM NaCl. Beads were washed with ten bed volumes of equilibration buffer and then two bed volumes of 30âmM Bis-Trisâ¢HCl pH 6.5, 350âmM NaCl at 4â°C. The Ub1â74~2-mercaptoethanesulfonate (MESNa) thioester was obtained by incubating the resin in 2 bed volumes of 30âmM Bis-Trisâ¢HCl pH 6.5, 350âmM NaCl, 200âmM MESNa at room temperature for 12â16âh. The cleaved protein was eluted and transferred to ice/water bath, then the resin was treated again with cleavage buffer for 12âh at 4â°C, followed by elution. Eluted fractions were combined, concentrated using 3âkDa MWCO Amicon filter (Milipore) to 8â10âmgâmlâ1 and treated with 1âM hydrazine at 30â°C for 30âmin. The resulting Ub1â74 hydrazide was separated by SEC using HiLoad 26/600 Superdex 75 prep grade column (GE Healthcare) equilibrated with 25âmM sodium phosphate pH 6.5, 350âmM NaCl at 4â°C. Fractions containing Ub1â74 hydrazide were pooled, concentrated, flash-frozen in liquid nitrogen, and stored at â80â°C.
To generate Ub1â74 acyl azide for coupling with H2N-GlyâPSAN, Ub1â74 hydrazide, at a concentration of 3âmM, was combined with 100âmM sodium citrate pH 3.0 on ice/water bath. NaNO2 was added to a final concentration of 50âmM (from 0.5âM stock adjusted to pH 5 with HCl on ice/water bath immediately before use) and the reaction transferred to â20â°C (ice:NaCl bath at 3:1 ratio) for 5âmin. An equal volume of 250âmM H2N-GlyâPSAN (HCl salt) in 1âM sodium HEPES pH 8 was added immediately after dissolving and the reaction incubated for 10âmin on ice/water bath followed by 10âmin at room temperature, and buffer exchanged into 20âmM sodium HEPES pH 8, 350âmM NaCl at 4â°C using a 7âkDa MWCO Zeba Spin Column (Thermo Fisher Scientific). Ub-PSAN was separated by SEC using Superdex 75 Increase 10/300 GL column (GE Healthcare) equlibrated in 20âmM sodium HEPES pH 8, 350âmM NaCl at 4â°C. The molecular weight of Ub-PSAN was confirmed by ultraperformance liquid chromatography electrospray mass spectrometry using Acquity UPLC-MS (Waters). Ub-PSAN was concentrated to ~1âmM, flash-frozen in liquid nitrogen, and stored at â80â°C.
Preparation of E1âUb(T)âE2 and E2âUb(T)âE3HECT complexes
To obtain E2âUb vinyl thioether for reactions with E1 and E3HECT, a mixture of 0.6âmM Ub-PSAN, 0.4âmM Ubc4(C21S/C107S), 20âmM sodium HEPES pH 8.0, 200âmM NaCl was incubated for 1âh at 4â°C. The E2âUb vinyl thioether product was separated by SEC on a HiLoad 26/600 Superdex 75 prep grade column (GE Healthcare) equilibrated in the same buffer.
To obtain E1âUb(T)âE2, a mixture of 75âμM E2âUb vinyl thioether, 50âμM Uba1 (lacking the first 12 amino acids), 20âmM sodium HEPES pH 8.0, 100âmM NaCl, 0.2âmM TCEP was incubated for 1âh at room temperature, and then incubated on ice/water bath. The 30âml total reaction volume was divided into four 7.5âml portions. Each portion was applied to a 5âml StrepTrap HP column (GE Healthcare) equilibrated in Trisâ¢HCl pH 7.2, 200âmM NaCl, 0.2âmM TCEP at 4â°C. After washing with 10 column volumes of equilibration buffer, proteins were eluted in the same buffer containing desthiobiotin at a final concentration of 2.5âmM. Eluted fractions from all runs were combined and incubated with TEV protease for 9âh at 4â°C to remove the StrepTagâTEV tag from Ub. After tag cleavage, E1âUb(T)âE2 was separated by anion-exchange chromatography using MonoQ 5/50 GL column (GE Healthcare) in 20âmM Trisâ¢HCl pH 7.2, 0.1âmM TCEP and a linear gradient of 50â400âmM NaCl at 4â°C. Fractions containing E1âUb(T)âE2 were concentrated to ~12âmgâmlâ1, flash-frozen in liquid nitrogen, and stored at â80â°C.
To obtain E2âUb(T)âE3HECT, a 30âml solution containing 75âμM E2âUb vinyl thioether, 75âμM Pub2HECT, 20âmM sodium HEPES pH 8.0, 200âmM NaCl, 0.5âmM TCEP was incubated for 8âh at room temperature, then moved to ice/water bath. The reaction mixture was subjected to StrepTag affinity chromatography and TEV protease cleavage as described above for E1âUb(T)âE2 complex. After cleavage, E2âUb(T)-E3HECT was isolated with two successive rounds of anion-exchange chromatography using a MonoQ 5/50 GL column (GE Healthcare) equilibrated with 20âmM Trisâ¢HCl pH 7.5, 0.1âmM TCEP with a linear gradient of 50â280âmM NaCl at 4â°C. Fractions containing E2âUb(T)âE3HECT were concentrated to ~13.5âmgâmlâ1, flash-frozen, and stored at â80â°C.
Cryo-EM sample preparation and data collection
Prior to grid preparation, an aliquot of E1âUb(T)âE2 or E2âUb(T)âE3HECT was rapidly thawed in a room temperature water bath and centrifuged for 10âmin at 4â°C at 18,000g. E1âUb(T)-E2 was diluted to 3âmgâmlâ1 in Trisâ¢HCl pH 7.2, 100âmM NaCl and preincubated for 30âmin on ice/water bath with 5âmM MgCl2, 1âmM ATP, and ~0.8 molar equivalents of Ub. E2âUb(T)âE3HECT was diluted to 4.5âmgâmlâ1 in Trisâ¢HCl pH 7.2, 100âmM NaCl. Prior to vitrification, CHAPSO was added to samples at final concentrations of 0.05% and 0.1% w/v for E1âUb(T)âE2 and E2âUb(T)âE3HECT complexes, respectively. Four microlitres of sample was applied to freshly glow-discharged UltrAuFoil 300 mesh R1.2/1.3 grids (Quantifoil) at 100% humidity at 25â°C. After 8âs, samples were blotted for 3.5â4.0âs and vitrified by plunging into liquid ethane using a Vitrobot Mark IV (FEI-Thermo Fisher).
Cryo-EM data were collected at MSK Richard Rifkind Center for Cryo-EM, using a Titan Krios transmission electron microscope (FEI-Thermo Fisher) operated at 300âkeV. Cryo-EM movies (40 frames per movie, 4âs exposure time) were recorded at a dose rate of ~20 eââpixelâ1âsâ1 using a K3 Summit direct electron detector (Gatan) operated in super-resolution mode at a physical pixel size of 1.064âà . Automated data collection was performed in Serial EM54 using image shift to record data from nine ice holes per stage movement. Two datasets for E1âUb(T)âE2 were obtained with 12,132 and 14,545 movies each, and 2 datasets for E2âUb(T)âE3HECT were obtained with 15,705 and 14,101 movies each.
Cryo-EM image processing
Initial data processing steps were similar for E1âUb(T)âE2 and E2âUb(T)âE3HECT. Movie frames from each session were gain normalized, 2à Fourier cropped, aligned and summed with and without dose-weighting using MotionCor255. Estimation of the contrast transfer function (CTF) was performed using Gctf56 from non-dose weighted micrographs. Micrographs with estimated resolution limits worse than 4.5âà , poor CTF fit or with crystalline ice were discarded. Initial particle sets were obtained by reference-free auto-picking with Laplacian-of-Gaussian filtering in RELION 3.157,58. Subsequent steps were performed in cryoSPARC 4.0.259 with the exception of particle picking using Topaz60 and Bayesian polishing performed in RELION 3.161. UCSF Pyem was used to convert particle metadata from cryoSPARC to RELION format62. Particles from each dataset underwent several rounds of 2D classification to remove junk particles and to obtain subsets from 2,000 random micrographs for training neural network-based particle picking in Topaz. Trained models were applied to full datasets, and identified particles were extracted using a 256-pixel box size.
For E1âUb(T)âE2, Topaz-picked particles were used in rounds of 2D classifications to remove junk particles. Two successive rounds of ab initio 3D reconstruction and heterogeneous refinement were performed, first with three classes, then with four, each time removing classes lacking secondary structure features. 1,826,497 particles were re-extracted using a 384-pixel box size and pooled for a single ab initio 3D reconstruction followed by non-uniform 3D refinement63, resulting in a consensus map with a nominal resolution of 3.0âà . To remove low-quality particles, four consecutive rounds of heterogeneous refinement were initialized with four copies of a consensus map lowpass filtered to 15âà and one copy lowpass filtered to 30âà . Particles assigned to the lowest resolution class were discarded after each round. Particles were recentred and subjected to non-uniform 3D refinement and Bayesian polishing, followed by 2D classification to remove images with artefacts resulting in 1,610,345 particles that yielded a consensus map with a nominal resolution of 2.64âà , which improved to 2.51âà after per-particle defocus refinement, per optics group estimation of the beam tilt, trefoil, spherical and fourth-order aberrations, followed by second round of per-particle defocus refinement. Particles were next subjected to heterogeneous refinement (with 6 classes and 2à binning) initialized with 20âà lowpass-filtered consensus map resulting in four classes with Ub(A) and two without Ub(A) that were combined to yield two particle sets for doubly and singly loaded complexes (1,295,595 and 314,750 particles respectively), which after non-uniform 3D refinement, resulted in consensus maps at nominal resolutions of 2.50 and 2.79âà , respectively. To resolve Ub(T) conformations in doubly and singly loaded complexes, particles were then subjected to 3D classification without image realignment (10 classes, target resolution 7âà , number of O-EM epochs 8, O-EM learning rate init 0.3, initialization mode principal components analysis (PCA), force hard classification on) with a mask focused on Ub(T) region. Maps were generated by 3D reconstruction using particles from each class and their alignment information and a half-set splits from the gold-standard refinement of their parental sets (consensus maps). The 3D classes used for model building were further subjected to non-uniform 3D refinement. The continuum of E1 SCCH rotation relative to E1 adenylation domains (IAD and AAD) was resolved by 3D variability analyses64 (3 modes to solve, filter resolution 5âà ) performed separately for particles corresponding to doubly and singly loaded complexes, using a mask encompassing the E1âE2 region but excluding Ub(T) regions. In both cases, particles were sorted into five clusters based on values of the latent coordinate of the component 1, best capturing rotation of E1 SCCH domain. Five clusters were chosen because this number adequately tracked SCCH movement (1.5° to 2° of rotation per cluster) while ensuring sufficient particles within each cluster for further analysis. Particles from each cluster were subjected to non-uniform refinement, resulting in maps with nominal resolutions of 2.95â3.16âà for singly loaded complexes and 2.67â2.86âà for doubly loaded complexes. Ub(T) conformations in each cluster were resolved through 3D classifications without image realignment (10 classes, target resolution 7âà , number of O-EM epochs 8, O-EM learning rate init 0.3, initialization mode PCA, force hard classification on) using a focus mask on the Ub(T) region. Maps were generated by 3D reconstruction using particles from each class and their alignment information and half-set splits from the gold-standard refinement of parental sets (clusters 1â5). The 3D classes used for model building were further subjected to non-uniform 3D refinement. Where applicable, maps were lowpass filtered to 5âà using a 10th-order Butterworth filter. Statistics for data collection are listed in Supplementary Table 1.
For E2âUb(T)âE3HECT, Topaz-picked particles were selected after 2D classification from each dataset and re-extracted using 320-pixel box size, combined, and subjected to rounds of 2D classification, resulting in 2,704,150 particles. Ab initio 3D reconstructions using six classes were performed, followed by heterogeneous refinement (with 2à binning). Particles from classes with defined secondary structure features (2,511,724 particles) were combined and used in single ab initio 3D reconstruction followed by non-uniform 3D refinement to a map with a nominal resolution of 3.30âà . Particles were recentered, subjected to non-uniform 3D refinement and Bayesian polishing, followed by 2D classification to remove images with artefacts, and per-particle defocus refinement, resulting in 2,428,313 particles that yielded a consensus map with a nominal resolution of 3.09âà . Subsequently, 3D variability analysis (3 modes to solve, filter resolution 5âà ) was performed with a mask encompassing the entire complex. Particles were then split into 20 clusters based on all solved modes of 3D variability, an empirically determined number that resulted in best separation between complexes. Each cluster was subjected to non-uniform 3D refinement, resulting in maps with nominal resolutions of 3.27â3.56âà . The 8 clusters resulted in maps with no apparent or poorly resolved Ub(T) density. Ub(T) densities were resolved in maps for 12 clusters with 6 maps capturing Ub(T) at positions proximal to E2 and 6 other maps with Ub(T) in positions more distal from E2 (states 2â7). All 12 maps revealed Ub(T) C-terminal residues between E2 and E3HECT. Particles from 6 clusters containing Ub(T) at a position most proximal to E2 (790,208 particles) were combined and used for non-uniform 3D refinement resulting in a map with a nominal resolution of 3.17âà . To select the best particles and to improve the map at the transthiolation active site, a 3D classification without image alignment (4 classes, target resolution 7âà , number of O-EM epochs 8, O-EM learning rate init 0.3, initialization mode PCA, force hard classification on) was performed with a mask focused on the transthiolation site. One of four classes showed improved density corresponding to the transthiolation site, including E2 amino acids around its active site. This class containing 204,763 particles was subjected to non-uniform 3D refinement to yield a map with a nominal resolution of 3.23âà (state 1). Reported resolutions were determined using the gold-standard 0.143 criterion based on Fourier shell correlation. Statistics for data collection are listed in Supplementary Table 1.
Model building and refinement
Initial coordinates were generated by docking individual chains from reference structures into cryo-EM maps in UCSF Chimera65 followed by manual building in Coot66. For E1âUb(T)âE2, the crystal structures of Uba1âUbc4/Ub/ATP·Mg and Ub (Protein Data Bank (PDB): 4II217 and 6O8215, respectively) were used. For E2âUb(T)âE3HECT, a homology model of Pub2HECT was obtained from SWISS-MODEL67 based on crystal structures of UbcH5Bâ¼Ub-NEDD4L (PDB: 3JW025), Ubc4 and Ub (PDB: 4II217 and 6O8215, respectively). Coordinates for all models were produced via iterative rounds of refinement and building in real space using Phenix and Coot66,68. Geometry restraints for the linker (transthiolation intermediate analogue) were generated using Phenix.elbow69. MolProbity was used to evaluate model integrity70. Structure representations were generated using UCSF ChimeraX71. 2D slice views of electron microscopy maps were visualized using IMOD 4.1172. Statistics for model refinement are listed in Supplementary Table 1.
Preparation of singly loaded fluorescein-Ub~E1 complex
To generate thioester-linked fluorescein-Ub~E1 complex, 125âμl reaction containing full-length Uba1 (8âμM), substochiometric amount of fluorescein-Ub (6âμM) in 20âmM sodium HEPES pH 7.5, 100âmM NaCl, 0.1âmM TCEP, 5âmM MgCl2 and 1âmM ATP was incubated at 10âmin at 30â°C. The mixture was then cooled to 4â°C and separated by anion-exchange chromatography using MonoQ 5/50 GL column (GE Healthcare) in 20 sodium HEPES pH 7.5, 0.1âmM TCEP and a linear gradient of 100â400âmM NaCl. SDSâPAGE analysis of purified fluorescein-Ub~E1 incubated with and without β-mercaptoethanol confirmed its sensitivity to reducing agent, consistent with a singly loaded fluorescein-Ub~E1 and without Ub bound non-covalently to the adenylation site.
E1âE2 single-turnover Ub thioester transfer assays
The Ub~AMP mimic (Ub-AMSN) was synthesized as described previously14,15,34. Single turnover assays were performed on ice/water bath. For each replicate, singly loaded fluorescein-Ub~E1 complex (10ânM) was freshly prepared and preincubated in 20âmM sodium HEPES pH 7.5, 100âmM NaCl, 0.1âmM TCEP, 0.1âmgâmlâ1 ovalbumin, either alone or with addition of ATP (4âmM), Ub and ATP (10âμM and 4âmM, respectively), Ub-AMSN (10âμM), or Ub-AMSN and PPi (10âμM and 4âmM, respectively) in 400âμl for 5âmin. A control without addition of Ubc4 (E2) was obtained by diluting a 100âμl aliquot with an equal volume of 20âmM sodium HEPES pH 7.5, 100âmM NaCl, 5âmM MgCl2, 0.1âmM TCEP, 0.1âmgâmlâ1 ovalbumin. Thioester transfer (chase) was initiated by diluting a 100âμl aliquot with an equal volume of 100ânM E2, 20âmM sodium HEPES pH 7.5, 100âmM NaCl, 5âmM MgCl2, 0.1âmM TCEP, 0.1âmgâmlâ1 ovalbumin. The resulting concentrations at time 0 were 5ânM for fluorescein-Ub~E1 complex and 50ânM for E2. Aliquots from indicated timepoints and a control without E2 were quenched by addition of LDS NuPAGE buffer supplemented with EDTA (final concentrations of 1à and 50âmM, respectively). Products were separated on 4â12% NuPAGE BIS-Tris gels with 1à MOPS running buffer (Thermo Fisher Scientific). To increase fluorescence signal by converting fluorescein to its di-anionic form, gels were incubated in 50âmM Trisâ¢HCl pH 9.5 for 3âmin prior to scanning using Amersham Typhoon 5 (Cytiva). Bands were quantified using ImageQuant software (Cytiva).
E1âE2 multiple turnover Ub thioester transfer assays
E1âE2 Ub thioester transfer assays were performed in 160âμl reactions with 1.5ânM full-length Uba1, 200ânM of indicated variant of Ubc4, 5âμM Ub, 20âmM sodium HEPES pH 7.5, 50âmM NaCl, 5âmM MgCl2, 0.1âmM TCEP. Reactions were conducted at 22â°C and initiated by adding ATP to a final concentration of 200âμM including a control that lacked ATP. Aliquots were removed at indicated timepoints and quenched by addition of LDS NuPAGE buffer supplemented with EDTA (final concentrations of 1à and 50âmM, respectively) and products were separated on 12% NuPAGE BIS-Tris gels with MOPS running buffer (Thermo Fisher Scientific). Gels were stained using Flamingo dye (Bio-Rad), scanned using Amersham Typhoon FLA 9500 and quantified using ImageQuant software (Cytiva).
E2~Ub(T) to E3HECT pulseâchase Ub thioester transfer assays
For E2~Ub(T) to E3HECT~Ub thioester transfer assays, for each replicate, indicated mutational variants or wild-type Ubc4 (7âμM) were charged with wild type or indicated mutational variant of fluorescein-Ub (12âμM) using E1 (300ânM) in 20âmM sodium HEPES pH 7.5, 100âmM NaCl, 5âmM MgCl2, 2âmM ATP, 0.1âmM TCEP in 40âμl reaction volume. After incubation for 30âmin at 25â°C, reactions were transferred to ice/water bath and E1 activity was quenched by fourfold dilution with 20âmM sodium HEPES pH 7.5, 100âmM NaCl, 40âmM EDTA. Subsequent steps were performed on ice/water bath. Mixtures were diluted with 20âmM sodium HEPES pH 7.5, 100âmM NaCl, 0.1âmM TCEP, 0.1âmgâmlâ1 ovalbumin to a final E2~Ub(T) concentration of 30ânM. A control without E3HECT was obtained by diluting a 100âμl aliquot with an equal volume of 20âmM sodium HEPES pH 7.5, 100âmM NaCl, 0.1âmM TCEP, and 0.1âmgâmlâ1 ovalbumin. Thioester transfer reactions (chase) were initiated by diluting a 100âμl aliquot with an equal volume of a solution of a specific variant of Pub2HECT at 40ânM in 20âmM sodium HEPES pH 7.5, 100âmM NaCl, 0.1âmM TCEP, and 0.1âmgâmlâ1 ovalbumin. Concentrations at time 0 were 15ânM for fluorescein-Ub~E2 and 20ânM for E3HECT. Aliquots (40âμl) were removed at indicated timepoints and quenched by addition of LDS NuPAGE buffer (final concentration of 1Ã). Products were separated on 4â12% NuPAGE BIS-Tris gels with MOPS running buffer (Thermo Fisher Scientific). To increase fluorescence signal by converting fluorescein to its di-anionic form, gels were incubated in 50âmM Trisâ¢HCl pH 9.5 for 3âmin prior to scanning using Amersham Typhoon 5 (Cytiva). Bands were quantified using ImageQuant software (Cytiva).
Statistics and reproducibility
Generation of the E1âUb(T)âE2 and E2âUb(T)âE3 transthiolation analogues for structural analysis was reproduced with at least three independent purifications. All biochemical experiments were replicated three times, reproduced with at least two independent purifications, and reproduced independently at least twice. Statistical analyses and graphing of the data were performed using Prism 10.1.0 (GraphPad Software). Number of replicates and details on statistical analyses and test are provided in the methods pertaining to each experiment and/or the appropriate legend.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.