Protein expression and purification
The full-length, codon-optimized genes encoding human RAGC with the S75N substitution and human RAGA with the Q66L substitution were synthesized (Twist Bioscience) and individually cloned into a pCAG vector. Mutations were introduced in the genes encoding RAGA (Q66L) and RAGC (S75N) to lock the active state of RAG GTPases (RAGAGTP–RAGCGDP). The RAGC (S75N) construct included sequence for a TEV-cleavable GST tag at the N terminus, whereas the RAGA (Q66L) construct was tagless. For expression and purification of RAG GTPases, HEK293F GnTI- cells were transfected with a total of 1 mg plasmid DNA (550 μg for RAGA and 450 μg for RAGC) and 4 mg polyethylenimine (PEI) (Sigma-Aldrich) per l at a density of 1.8 × 106 cells per ml. Cells were collected after 72 h and lysed by gentle nutation in wash buffer (50 mM HEPES, 150 mM NaCl, 2.5 mM MgCl2, 1 mM TCEP, pH 7.4) supplemented with 0.4% CHAPS and Protease Inhibitor (Roche) for 1 h. The lysate was cleared by centrifugation at 35,000g for 35 min. The supernatant was incubated with glutathione Sepharose 4B (GE Healthcare) resin for 2 h. The resin was then washed first with a modified wash buffer (200 mM NaCl and 0.3% CHAPS) and then with wash buffer. The complex was eluted by on-column TEV cleavage overnight without nutation. Eluted complexes were concentrated and further purified by size exclusion chromatography (SEC) using a Superose 200 10/300 GL (GE Healthcare) column equilibrated with wash buffer.
The full-length, codon-optimized genes encoding the human Ragulator complex (LAMTOR1–LAMTOR5) were synthesized (Twist Bioscience) and individually cloned into a pCAG vector. The Lamtor1 construct includes sequence for a TEV-cleavable GST tag at the N terminus, followed by sequence for a His6 tag, which replaces its first seven residues. The Lamtor2 construct features sequence for a TEV-cleavable tandem 2× Strep II-1× Flag tag (2SF-TEV) at the N terminus. A total of 1 mg plasmid DNA (200 μg each of the five constructs) and 4 mg PEI (Sigma-Aldrich) were used to transfect HEK293F GnTI- cells at a density of 1.8 × 106 cells per ml per l. The cells were pelleted after 72 h of transfection and lysed in wash buffer containing 1% Triton X-100 and protease inhibitor. The cleared supernatant after centrifugation was applied to glutathione Sepharose 4B (GE Healthcare) resin and incubated for 2 h. The complex was eluted by on-column TEV cleavage overnight without nutation and supplemented with a final concentration of 0.5 mM EDTA. Further purification was performed by SEC using a Superdex 200 10/300 GL column equilibrated with wash buffer. The fractions containing all five subunits were pooled and concentrated.
To assemble the RAG–Ragulator complex, an excess amount of RAG was incubated with Ragulator and Ni-NTA resin (Thermo Scientific) at 4 °C for 1 h. Excess RAG was removed by washing the resin with wash buffer containing 40 mM imidazole. The assembled RAG–Ragulator complex was eluted using wash buffer with 250 mM imidazole and further purified by SEC with a Superdex 200 10/300 GL column.
The human mTORC1 complex was purified in a manner similar to that of previous methods20. The full-length, codon-optimized genes encoding human mTOR, RAPTOR and MLST8 were cloned into pCAG vectors (mTOR with TEV-cleavable 2SF, RAPTOR with uncleavable 2SF and MLST8 with uncleavable 2SF). A total of 1.35 mg plasmid DNA (900 μg for mTOR, 250 μg for RAPTOR and 200 μg for MLST8) and 4 mg PEI (Sigma-Aldrich) were used to transfect HEK293F GnTI- cells at a density of 1.8 × 106 cells per ml per l. Cells were collected 72 h after transfection and lysed in the same buffer used for RAGA–RAGC purification. The complex was purified using Strep-Tactin resin (IBA Lifesciences) and eluted with wash buffer containing 10 mM d-desthiobiotin. The eluate was then diluted in an equal volume of salt-free buffer (50 mM HEPES, 1 mM TCEP, pH 7.4) and applied to a 1-ml HiTrap Q column (GE Healthcare). The mTORC1 complex and free RAPTOR were separated with a 20-ml salt gradient to a final concentration of 0.5 M NaCl, using salt-free buffer and high-salt buffer (50 mM HEPES, 1 M NaCl, 1 mM TCEP, pH 7.4). The flow rate was 0.2 ml min−1, and the fraction size was 0.2 ml. The fractions containing the mTORC1 complex and free RAPTOR were collected and concentrated with Amicon Ultra-4 concentrators. Mutations in the genes encoding mTOR and Raptor were generated using NEBuilder HiFi DNA Assembly. The mTORC1 complex mutants were produced by altering the combination of wild-type and mutated genes during transfection and purified as described above.
Plasmids containing full-length genes encoding human 4E-BP1 and RHEB were gifts from the Zoncu laboratory (University of California, Berkeley). These genes were individually cloned into a 2GT vector from QB3 MacroLab (https://qb3.berkeley.edu/facility/qb3-macrolab/), which features sequence for a TEV-cleavable tandem GST–His6 tag at the N terminus. Genes encoding 4E-BP1 and RHEB were overexpressed in Escherichia coli Rosetta 2(DE3) strains and purified using the same method. E. coli cells were grown in LB medium at 37 °C until an OD of 0.6 was reached and then induced by adding 0.2 mM IPTG at 18 °C overnight. Cells were collected, resuspended in Ni buffer (50 mM Tris-Cl, pH 7.5, 300 mM NaCl, 20 mM imidazole, 5 mM 2-mercaptoethanol, 1 mM PMSF) and lysed by sonication. Protein was purified using HisPur Ni-NTA Resin (Thermo Scientific), washed with Ni buffer containing 40 mM imidazole and eluted with 250 mM imidazole. The GST–His6 tag was cleaved by incubating with TEV enzyme overnight. Further purification was performed by SEC using a Superdex 75 10/300 GL column equilibrated with buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM TCEP). Fractions containing the desired proteins were passed through the Ni column to remove residual GST–His6 and then concentrated.
To charge RHEB with GTPγS (Abcam) or GDP (Sigma-Aldrich), RHEB was first diluted in buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM TCEP, 5 mM EDTA) and then supplemented with GDP or GTPγS at a 30-fold molar excess. The mixture was incubated at 30 °C for 1 h, followed by adding 20 mM MgCl2. Further purification was carried out by SEC using a Superose 75 10/300 GL (GE Healthcare) column to remove excess nucleotides.
All steps of protein purification were performed at 4 °C, and aliquoted proteins were flash frozen in liquid nitrogen and stored at −80 °C.
LUV preparation
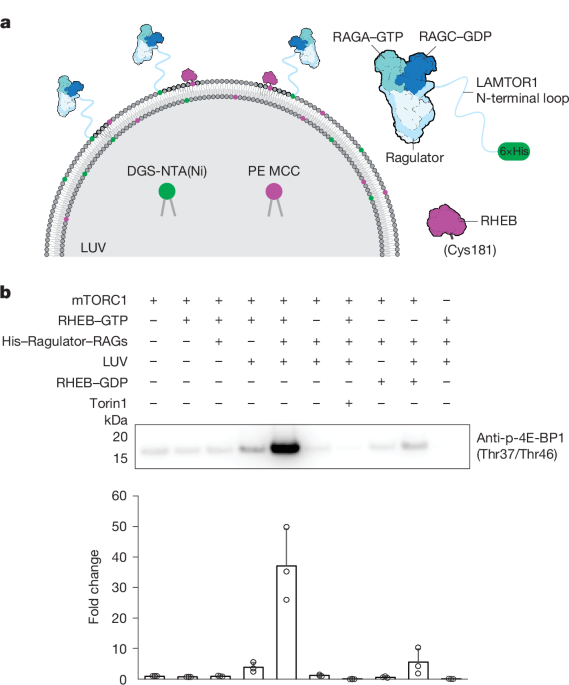
A lipid mixture was prepared in a glass vial using the lipid composition shown in Supplementary Table 1. To form a thin film on the glass wall, the glass vial was slowly shaken on a vortex while drying under nitrogen gas. The glass vial was then placed in a vacuum oven overnight at room temperature to evaporate any remaining solvent. Lipids were hydrated in a lipid buffer (25 mM HEPES, pH 7.2, 100 mM NaCl) to a final concentration of 1.8 mM for 1 h, with intermittent vortexing during hydration. The solution was transferred to a 15-ml Eppendorf tube and subjected to nine freeze–thaw cycles using liquid nitrogen and a 40 °C water bath. The lipid mixture was either stored at −80 °C or immediately extruded using an Avanti Polar Lipids Mini Extruder (610023) at least 41 times through a 200-nm filter (Whatman Nuclepore Track-Etched Membranes, diameter of 19 mm) for mTORC1 kinase activity assays and cryo-EM studies. Filters of different diameters were also used to generate liposomes of various sizes to assess the effect of liposome size on mTORC1 kinase activity. The mean diameter of extruded liposomes was measured using dynamic light scattering (Zetasizer Ultra). The average sizes of liposomes for filters of 400 nm, 200 nm, 100 nm, 50 nm and 30 nm were about 355 nm, 148 nm, 110 nm, 81 nm and 67 nm, respectively. The lipid solution after extrusion was stored at 4 °C for up to 2 weeks.
mTORC1 kinase activity with LUVs
The kinase assay was conducted in a buffer containing 25 mM HEPES (pH 7.2), 100 mM NaCl, 10 mM imidazole, 10 mM MgCl2 and 2 mM DTT at 30 °C for 10 min in a final volume of 50 μl. First, RHEB was incubated with liposomes at concentrations of 0.25 μM and 0.18 mM, respectively, at 4 °C overnight in 40 μl lipid buffer. In parallel, RHEB alone and liposomes alone were diluted to the same concentrations in lipid buffer and incubated at 4 °C overnight. After overnight incubation, the reactions were stopped with 2 mM DTT at room temperature for 30 min, followed by adding 10 mM MgCl2 and 10 mM imidazole. The His6-tagged RAG–Ragulator complex was then added to the reactions and incubated on ice for 30 min. Subsequently, 4E-BP1 and mTORC1 were added to the reactions at final concentrations of 10 μM and 5 nM, respectively. The reactions were initiated by adding ATP at a final concentration of 1 mM and incubated in a thermocycler (Bio-Rad, T100) at 30 °C for 10 min. The reactions were stopped by diluting tenfold into a urea denaturing buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 8 M urea). All reactions were then diluted into 4× NuPAGE LDS sample buffer and boiled for 2 min. Proteins were resolved on a 4–12% NuPAGE Bis-Tris gel and transferred to PVDF membranes using the Trans-Blot Turbo Transfer System. Western blotting was performed using the anti-p-4E-BP1 antibody (Cell Signaling Technology, 2855, 1:10,000 dilution). For dot blot analysis, 2 μl of the denatured sample in urea buffer was directly applied to nitrocellulose membranes, and protein was detected with the same 4E-BP1 antibody. Unprocessed images are included in Supplementary Fig. 1.
Cryo-EM sample preparation and imaging
The mTORC1–RHEB–RAG–Ragulator–4E-BP1 complex on liposomes was reconstituted with the following steps. First, a mixture of RHEB and liposomes (200 nm) was incubated at 4 °C overnight in lipid buffer at concentrations of 8 μM and 1.8 mM, respectively. The RHEB–liposome mixture was supplemented with 2 mM DTT and 10 mM MgCl2 and incubated at room temperature for 30 min. In parallel, mTORC1 was incubated with His6–RAG–Ragulator in lipid buffer containing 5 mM TCEP at concentrations of 1 μM and 4 μM, respectively. Equal volumes of the RHEB–liposome mixture and the mTORC1–RAG–Ragulator mixture were then combined and incubated on ice for 2 h. Finally, 4 μM of 4E-BP1 and 1 mM of AMPPNP were added to the mixture for 10 min before application to cryo-EM grids for vitrification.
Cryo-EM samples were prepared by applying 3 μl of the aforementioned complex to a glow-discharged (PELCO easiGlow, 45 s in air at 15 mA and 0.37 mbar) holey carbon grid (C-flat, 2/1-3C-T) and vitrified using the FEI Vitrobot Mark IV System (Thermo Fisher Scientific). The samples were incubated on grids for 1 min and blotted for 3 s with a blot force of 15, using two Whatman 595 papers on the sample side and one Whatman 595 paper on the backside, at 6 °C with 95% relative humidity.
Cryo-EM images of the mTORC1–RHEB–RAG–Ragulator–4E-BP1 complex on liposomes were recorded using a Titan Krios G3 microscope (Thermo Fisher Scientific) equipped with a Gatan Quantum energy filter (slit width of 20 eV) and operated at 300 kV. Automated data acquisition was performed using SerialEM50 on a K3 Summit direct detection camera (Gatan) in superresolution correlated double-sampling mode with a pixel size of 0.52 Å and a defocus range of −0.9 to −2.2 μm. A total of 36 exposures per stage shift were enabled by large beam shift. Beam intensity was adjusted to a dose rate of around 1 e− Å−2 per frame for a 30-frame movie stack with a total exposure time of 5.4 s. A total of 58,092 movies were recorded.
Cryo-EM data processing
The data-processing scheme for the mTORC1–RHEB–RAG–Ragulator–4E-BP1 complex on liposomes using cryoSPARC (v.4)51 is shown in Extended Data Fig. 1, and statistics are summarized in Table 1. Owing to the uneven distribution of liposomes in grid squares, 29,301 micrographs containing liposomes were manually selected for processing. Owing to the size of the dataset, micrographs were split and processed following the same protocol and then combined at the homogeneous refinement stage. Initially, the Blob Picker was used to maximize the number of particles. Two-dimensional classification was only used to remove obvious junk particles (for example, ice and chaperonin contaminants). An initial model of mTORC1 from an ab initio reconstruction in a previous dataset and three bad initial models from ab initio reconstruction in this dataset were used for heterogeneous refinement. Iterative heterogeneous refinement was performed to select good particles, retaining rare views that may not have been identified in 2D classification. To minimize the possibility of membrane density biasing the alignment in the first iterations of refinement, the original initial models were used instead of reconstructions from each heterogeneous refinement. After extensive cleaning using heterogeneous refinement, particles were merged, and duplicates were removed with a cutoff distance of 50 Å. Additional heterogeneous refinement was used to further sort out particles. Homogeneous refinement was then performed for the full dataset. After identifying a good particle set, it was used to train Topaz particle picking52. Particles from Topaz picking underwent the same sorting procedure and were merged with the blob-picked particles, with duplicates removed using a cutoff distance of 50 Å.
In total, 337,347 particles containing protein complexes and membranes were selected, and reference-based motion correction was used to produce polished particles53. Symmetry expansion, particle subtraction and local refinement were used to produce an overall cryo-EM map of an asymmetric unit. Focused 3D classification was then used to separate particles without extra RAG–Ragulator copies and with either one or two extra copies of RAG–Ragulator. Further particle subtraction and local refinement were used to focus on either mTOR–RHEB–MLST8 or RAPTOR–RAG–Ragulator subcomplexes. Focused 3D classification was used to separate the intermediate and fully active states of mTOR. Populations containing extra copies of RAG–Ragulator were pooled together, and further particle subtraction and local refinement were used to obtain the cryo-EM map of MLST8–RAG–Ragulator.
In summary, 179,506 particles were refined to 3.23 Å with C2 symmetry for the mTORC1–RHEB–RAG–Ragulator–4E-BP1 complex without extra RAG–Ragulator copies on the membrane. Local refinement of mTOR–RHEB–MLST8 (359,012 particles after symmetry expansion) and RAPTOR–RAG–Ragulator (189,975 particles after symmetry expansion) yielded maps of 3.12 Å and 2.98 Å, respectively. Further classification of mTOR–RHEB–MLST8 identified the fully active and intermediate conformations. Final refinement of mTOR–RHEB–MLST8 in fully active (133,193 particles after symmetry expansion) and intermediate (109,105 particles after symmetry expansion) conformations resulted in resolutions of 3.16 Å and 3.61 Å, respectively. Final resolutions for the mTORC1–RHEB–RAG–Ragulator–4E-BP1 complex containing either one (128,189 particles) or two (29,652 particles) extra copies of RAG–Ragulator were 3.47 Å (C1 symmetry) and 3.81 Å (C2 symmetry), respectively. Three-dimensional variability analysis for populations without extra RAG–Ragulator copies was accomplished with cryoSPARC v.4 (Extended Data Fig. 9).
The overall resolution of all these reconstructed maps was assessed using the gold-standard criterion of Fourier shell correlation54 at a cutoff of 0.143 (ref. 55). cryoSPARC v.4 was used to estimate local resolution.
Atomic model building and refinement
To build the atomic model for the mTORC1–RHEB–RAG–Ragulator–4E-BP1 complex on the membrane, we first fit our previous models into the cryo-EM map as a rigid body using UCSF ChimeraX56, with substituted models of mTOR and RAPTOR from AlphaFold2 prediction57. A composite map combining the local refinement maps was assembled in UCSF ChimeraX. Model refinement against local maps was accomplished using PHENIX for real-space refinement58. Manual model building was conducted with Coot59 and ISOLDE60 to iteratively inspect and improve local fitting. All figures were created using UCSF ChimeraX.
Cell culture
Inducible RAPTOR knockout MEFs were kindly provided by M.N. Hall (University of Basel). MEFs were cultured in DMEM High Glucose medium (ECM0728L, Euroclone) supplemented with 10% inactivated FBS (ECS0180L, Euroclone), 2 mM glutamine (ECB3000D, Euroclone), penicillin (100 IU ml−1) and streptomycin (100 μg ml−1) (ECB3001D, Euroclone) and maintained at 37 °C with 5% CO2. All RAPTOR mutants used in these cellular assays were generated using the QuikChange II-E Site-Directed Mutagenesis Kit (200555, Agilent Technologies). Cells were transfected in 10-cm dishes using Lipofectamine 2000 Transfection Reagent (Invitrogen).
Cell treatment
For experiments involving amino acid starvation, cells were rinsed twice with PBS and incubated for 60 min in amino acid-free DMEM (MBS6120661) supplemented with 10% dialysed FBS. Serum was dialysed against 1× PBS through dialysis tubing (molecular weight cutoff of 3,500 Da) to ensure the absence of contaminating amino acids. For amino acid refeeding, cells were restimulated for 30 min with a 1× water-solubilized mix of essential (11130036, Thermo Fisher Scientific) and non-essential (11140035, Thermo Fisher Scientific) amino acids resuspended in amino-acid-free DMEM supplemented with 10% dialysed FBS plus glutamine.
Western blotting
Antibodies used in cellular studies include anti-p-p70 S6K (Thr389) (1A5) (mouse mAb, 9206, 1:1,000 for western blotting), anti-p70 S6K (rabbit, 9202, 1:1,000 for western blotting), anti-4E-BP1 (rabbit, 9644, 1:1,000 for western blotting), anti-p-4E-BP1 (Ser65) (rabbit, 9456, 1:1,000 for western blotting) and anti-RAPTOR (24C12) (rabbit, 2280, 1:1,000 for western blotting) from Cell Signaling Technology; anti-GAPDH (6C5) (rabbit, sc-32233, 1:15,000 for western blotting) from Santa Cruz; and anti-Flag M2 (mouse, F1804, 1:1,000 for western blotting) from Sigma-Aldrich. Cells were rinsed once with PBS and lysed in ice-cold lysis buffer (250 mM NaCl, 1% Triton, 25 mM HEPES, pH 7.4) supplemented with protease and phosphatase inhibitors. Total lysates were passed ten times through a 25-gauge needle with a syringe, kept at 4 °C for 10 min and then cleared by centrifugation in a microcentrifuge (14,000 rpm at 4 °C for 10 min). Protein concentration was measured with the Bradford assay. We performed densitometry analysis to calculate the intensity of phosphorylated and total protein using ImageJ software. The ratios between the values of phosphorylated and total protein were normalized to those of a control condition. Values in quantitative graphs are mean ± s.e.m. of at least three independent experiments. For statistical analysis, two-way ANOVA and Dunnett’s post hoc test were used to compare differences between groups that had been split into two factors. Unprocessed gel images are shown in Supplementary Fig. 1.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.