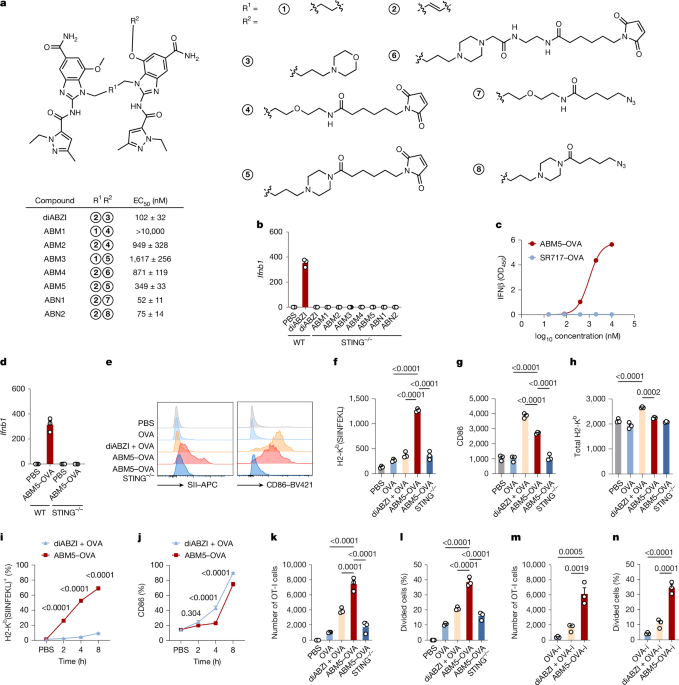
Synthesis of the SABER family
The interaction diagram of compound 2 and STING was analysed by Ligplot+ software (v2.2). Detailed synthetic procedures for the SABER family have been provided in the Supplementary Information. In brief, methyl-4-chloro-3-methoxy-5-nitrobenzene carboxylate was suspended in saturated ammonia and stirred at room temperature for 24 h, followed by heating at 50 °C for 2 h to obtain compound 1. Compound 1 was dissolved in dichloromethane (DCM), then boron bromide was injected dropwise, and stirred at room temperature for 16 h. The result solution was poured into ice water and stirred vigorously for 30 min to obtain compound 6. Compound 6 was dissolved in N,N-dimethylformamide (DMF). Then, tert-butyl 4-(3-(tolylenesulfonyloxy) propyl) piperazine-1-carboxylate and N,N-diisopropyl-ethylamine (DIPEA) were added and stirred at 80 °C for 4 h to obtain compound 7. The parallel synthesis process was initiated by dissolving compound 1 in DMF, followed by adding tert-butyl (4-aminobut-2-en-1-yl) carbamate and DIPEA and heating at 120 °C for 16 h. The crude solution was dissolved in DCM, and stirred with trifluoroacetic acid (TFA) to obtain compound 4. The dried compounds 4 and 7 were dissolved in isopropanol, and heated at 120 °C for 21 h with DIPEA. The crude solution was cooled. The brick-red precipitation was collected, dissolved in methanol with sodium dithionite, stirred at room temperature for 15 min, and quenched by adding sodium bicarbonate to obtain compound 15. Compound 15 was dissolved in DMF, followed by dropwise injection of 1-ethyl-3-methyl-1H-pyrazole-5-carbonyl isothiocyanate (in 1,4-dioxane) in the ice bath, and stirred at 0 °C for 30 min. Then, N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDCI) and DIPEA were added and stirred overnight at room temperature to obtain compound 20. Compound 20 was dissolved in DMF and stirred at room temperature after adding TFA for 2 h. The crude product was then dissolved in DMF. N-succinimidyl 6-maleimidohexanoate and DIPEA were added to the solution in the ice bath. The mixture was slowly warmed up to room temperature with stirring for 1 h to obtain ABM5. Compounds were purified by column chromatography, qualified and quantified by nuclear magnetic resonance (Avance III, Bruker) and mass spectrometry (Orbitrap Exploris 480, Thermo Fisher).
All peptides including OVA (CSGLEQLESIINFEKL), OVA-i (CSGLEQLESIINFEKI), OVA–biotin (CSGLEQLESIINFEKbiotinL), OVA30aa (CSGLEQLESIINFEKLTEWTSSNVMEERKIK), SNT (CKDGIIWVATEGALNTPKDHIGT), Adpgk (CELASMTNMELM) and M27 (propargylglycine-REGVELCPGNKYEM) were synthesized by Syneptide. To obtain SABER–peptide conjugates, ABM5 was covalently coupled with cysteine added to the N terminus of peptides. ABM5 in DMF was mixed with peptides in DMSO at a molar ratio of 1.1. Then, an equal volume of PBS was added. The mixture was shaken at room temperature for 24 h, followed by adding 10 times equal molar of DIPEA, and shaken at room temperature overnight. For the disulfide-containing counterpart, ABD in DMF was mixed with peptides in DMSO containing 4.8% acetic acid and incubated overnight at 37 °C. To covalently conjugate M27 with ABN2, ABN2 in DMSO was mixed with M27 in DMSO at a molar ratio of 2. Then, 8.8 times equal molar of CuSO4 (aqueous) and 17.6 times equal molar of sodium ascorbate (aqueous) were added, and shaken at room temperature for 1 h. To be labelled by Cy5.5, peptides or SABER–peptides in DMSO were mixed with Cy5.5–NHS in DMSO and DIPEA at molar ratios of 1 and 2, respectively. The mixture was shaken overnight at room temperature. All crude products obtained above were purified by high-performance liquid chromatography (HPLC; 1260, Agilent) and freeze-dried (FD-1A-50F+, Biocool). The molecular weight was confirmed by mass spectrometry.
To investigate the stability of conjugates, 10 μM ABM5–OVA or ABD–S–OVA were incubated with 1 mM or 10 mM GSH in PBS at 37 °C for 5 min, 1, 4 and 24 h, and then analysed by HPLC at 321 nm. The cleavage products were further confirmed by mass spectrometry.
Cell lines
STING−/− HeLa cells were provided by Z. Jiang at Peking University26. E.G7-OVA (CRL-2113) and Vero-E6 (CRL-1586) cells were obtained from the American Type Culture Collection. hACE2 stable expressing the HEK293T cell line (hACE2-293T) was obtained from PackGene Biotech. HeLa, THP-1, HEK293T, MC38 and B16F10 cell lines were maintained in our laboratory and validated by the STR identity. To generate B16-OVA cells, lentiviruses coding OVA were made by co-transfection of HEK293T cells with pCDH-ovalbumin-puro, psPAX2 and pMD2G plasmids. B16F10 cells were infected by the lentivirus and screened for single clones expressing OVA in a medium containing 2 μg ml−1 puromycin (MB2005, Meilunbio). The expression of OVA was validated by western blot. Only monoclonal cell lines that could express OVA over 15 generations were maintained and used for the following experiments. All cells were tested for Mycoplasma by a PCR kit (KP213-01, Tiangen).
Mice
C57BL/6 mice were obtained from GemPharmatech or the animal facility of Sun Yat-sen University. OT-I CD45.1+ transgenic mice were provided by C. Y. Yang at Sun Yat-sen University. STING−/− mice were provided by H. Yin at Tsinghua University, and were originally generated by Z. Jiang at Peking University. Batf3−/− mice were obtained from The Jackson Laboratory. Mice were housed in a specific pathogen-free environment in the animal facility of Sun Yat-sen University under 12-h light–dark cycles with ambient temperature between 21 and 24 °C and humidity between 40 and 70%. All the animal care and experimental procedures were performed with ethical compliance and approval by the IEC for Clinical Research and Animal Trials of the First Affiliated Hospital of Sun Yat-sen University or the Institutional Animal Care and Use Committee of Sun Yat-sen University. Male human ACE2-Tg (hACE2-Tg, CAG-hACE2-IRES-Luc-Tg) were purchased from the Shanghai Model Organisms Center, and housed in the animal facility of the School of Basic Medical Sciences at Fudan University. The protective effectiveness evaluation of vaccination in the mouse model of BA.5.2 infection was approved by the Laboratory Animal Ethics Committee of the School of Basic Medical Sciences at Fudan University.
Viruses
SARS-CoV-2 BA.5.2 variant was obtained from Shanghai Medical College, Fudan University, and the strain has been verified by next-generation sequencing twice. A plaque assay was used to quantify the viral titres using Vero-E6 cells. Experiments related to authentic SARS-CoV-2 were conducted in the BSL-3 Laboratory of Fudan University.
In vitro DC culture
Bone marrow cells were obtained from tibiaes and femurs of 4–6-week-old C57BL/6 mice. BMDCs were cultured as previously described7. In brief, bone marrow cells were cultured with 20 ng ml−1 of mouse granulocyte–macrophage colony-stimulating factor (GM-CSF; 576306, BioLegend) at the concentration of 1 × 106 per millilitre in complete RPMI-1640 (supplemented with 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 10% FBS) for 7 days at 37 °C and 5% CO2. FLT3L BMDCs were cultured as previously described49. In brief, bone marrow cells were resuspended at 2 × 106 per millilitre in complete RPMI-1640 containing 200 ng ml−1 recombinant FLT3L (550602, BioLegend) for 9 days at 37 °C and 5% CO2. THP-1-derived DC-like cells were cultured as previously described50. In brief, 2 × 105 per millilitre THP-1 cells were cultured in complete RPMI-1640 supplemented 0.05 mM 2-mercaptoethanol, 100 ng ml−1 human recombinant GM-CSF (572902, BioLegend) and 100 ng ml−1 human recombinant IL-4 (574002, BioLegend). The cells were cultured for 5 days at 37 °C in 5% CO2, with a media change on day 3.
EC50 measurement
The EC50 was determined by IFNβ expression in BMDCs. In brief, 1.5 × 104 BMDCs were seeded in 96-well plates and stimulated with SABERs, diABZI (tlrl-diabzi, InvivoGen), conjugates or their LNP-encapsulated counterparts at various concentrations. ABMs and ABNs were first blocked with cysteine or alkynylglycine plus ‘click’ reagents, respectively. After 24 h of inoculation, the supernatant of cells was collected for detecting IFNβ by an ELISA kit (439404, BioLegend). The EC50 was calculated from an agonist versus response curve by GraphPad Prism (v9.5.1).
Cytokine mRNA detection
Total RNA from BMDCs, HeLa, THP-1 or THP-1-derived DC-like cells was purified using the RNA Preparation Kit (AP-MN-MS-RNA-250, Axygen). cDNA synthesis was performed using the Thermo Scientific RevertAid RT Kit (K1691, Thermo Fisher). Real-time PCR was performed on the QuantStudio Real-Time PCR Detection System (Thermo Fisher) using the TB Green Master Mix (RR820Q, TaKaRa). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as an internal control. The primers for the detection of cytokine mRNA in BMDCs were: Cxcl10 forward: CCAAGTGCTGCCGTCATTTTC; Cxcl10 reverse: TCCCTATGGCCCTCATTCTCA; Gapdh forward: ATCAAGAAGGTGGTGAAGCA; Gapdh reverse: AGACAACCTGGTCCTCAGTGT; Ifnb1 forward: AGCTCCAAGAAAGGACGAACA; Ifnb1 reverse: GCCCTGTAGGTGAGGTTGAT.
The primers for detection of cytokine mRNA in HeLa, THP-1 or THP-1-derived DC-like cells were:
Cxcl10 forward: GTGGCATTCAAGGAGTACCTC; Cxcl10 reverse: TGATGGCCTTCGATTCTGGATT; Gapdh forward: GGAGCGAGATCCCTCCAAAAT; Gapdh reverse: GGCTGTTGTCATACTTCTCATGG; Ifnb1 forward: GCTTGGATTCCTACAAAGAAGCA; Ifnb1 reverse: ATAGATGGTCAATGCGGCGTC.
Immunofluorescence
For the colocalization of peptides, STING and various organelles, 1 × 105 HeLa or STING−/− HeLa cells were transfected with pCMV-STING1(human)-3×FLAG-WPRE-Neo in the presence of 10 μM cGAS inhibitor G140 (HY-133916, MCE) 24 h before the stimulation. BMDCs were seeded as mentioned above. The cells were treated with 5 μM biotin-labelled molecules, 2.5 μM Cy5.5-labelled molecules or 1 μM LNP-encapsulated Cy5.5-labelled molecules for the indicated time, fixed with 4% paraformaldehyde, and permeabilized by a permeabilization buffer containing saponin (P0095, Beyotime) with 30% FBS or normal goat serum. Samples were incubated with anti-FLAG (1:100; F1804, Sigma-Aldrich), anti-calreticulin (1:100; ab2907, Abcam), anti-TAP1 (1:20; 11114-1-AP, Proteintech), anti-proteasome 20S (1:100; ab22673, Abcam), anti-GM130 (1:100; 618022, BD) or anti-STING (1:100; MABF270, Sigma-Aldrich) antibodies at 4 °C overnight. Cells were washed and incubated with secondary antibodies at room temperature for 1 h, including AF488-conjugated anti-mouse IgG (1:300; 4408, CST), AF555-conjugated anti-rabbit IgG (1:300; ab150074, Abcam), STAR-ORANGE anti-mouse IgG (1:100; STORANGE-1001, Abberior) and STAR-RED anti-rabbit IgG (1:100; STRED-1002, Abberior). Cells were subsequently stained with DAPI (1:500; D9542, Sigma Aldrich) for 10 min at room temperature. The stained samples were mounted by ProLong Gold Antifade Mountant (P36982, Thermo Fisher) and subjected to imaging with confocal fluorescence microscopy (FV-3000, Olympus) or STED (STEDYCON, Abberior). FV31S-SW and STEDYCON Gallery software were used to collect and analyse imaging data. Mander’s colocalization coefficiency was calculated by JACoP plugin of ImageJ51.
TEM
To visualize cellular ultrastructure after SABER stimulation, 1 × 105 BMDCs were seeded in 12-well plates. Cells were incubated with 2.5 μM of diABZI or ABM5–OVA for 2 h, with PBS-treated cells serving as the negative control. Cells were fixed with 2.5% glutaraldehyde in PBS for 15 min at room temperature. Subsequently, the cells were stained with OsO4–potassium ferrocyanide, dehydrated and embedded in Epon. The ultrathin sections were obtained and examined by TEM (Talos L120C G2, Thermo Scientific).
Antigen cross-presentation and maturation of DCs
To evaluate antigen cross-presentation and STING activation-mediated upregulation of co-stimulatory factors, 4 × 105 BMDCs, STING−/− BMDCs or FLT3L BMDCs were pulsed by either peptides with or without diABZI or various conjugates for the indicated time. For inhibitor studies, various inhibitors, including MG132 (10 μM; HY-13259, MCE), PD150606 (10 μM; 26066, MCE), ERAP1-IN-1 (100 μM; HY-133125, MCE), diABZI (10 μM), H151 (10 μM; T5674, TargetMol) or imiquimod (10 μM; HY-B0180, MCE) were used to treat BMDCs 40 min before addition of ABM5–OVA, except brefeldin A (10 μg ml−1; 420601, BioLegend), which was added 2 h ahead. Cells were then pulsed with 0.5 μM ABM5–OVA for 8 h. BMDCs were then harvested and stained by FITC–anti-CD11c (1:100; 117306, BioLegend), APC–anti-H2-Kb-SIINFEKL (1:100; 141606, BioLegend), BV421–anti-CD86 (1:100; 105032, BioLegend), PE–anti-H2-Kb (1:100; 116508, BioLegend) and Live/Dead dye (1:400; 423107, BioLegend). In FLT3L BMDCs, cDC1s and cDC2s were identified by FITC–anti-CD11c (1:100; 117306, BioLegend), APC–anti-CD24 (1:100; 138505, BioLegend), BV421–anti-CD86 (1:100; 105032, BioLegend), PE/Cy7–anti-B220 (1:100; 103222, BioLegend), PerCP/Cy5.5–anti-CD172α (1:100; 144010, BioLegend), PE–anti-H2-Kb-SIINFEKL (1:100; 141603, BioLegend) and Live/Dead dye (1:400; 423107, BioLegend). The measurement of in vivo antigen cross-presentation and related DC subtypes was performed as in previous studies31,52. Mice receiving subcutatneous injection of 10 nmol LNP-encapsulated ABM5–OVA were euthanized 24 h later, and then draining lymph nodes were collected. Single cells were obtained by passing the tissues through 40-μm cell strainers, lysed with ACK lysis buffer (BL503A, Biosharp), blocked by anti-CD16/CD32 (1:400; 101301, BioLegend), and stained with FITC–anti-CD11c (1:100; 117306, BioLegend), BV711–anti-CD8a (1:100; 100747, BioLegend), APC/Cy7–anti-CD103 (1:100; 121431, BioLegend), PE–anti-CD11b (1:100; 101207, BioLegend), PB–anti-CD86 (1:100; 105021, BioLegend) and PE/Cy7–anti-H2-Kb-SIINFEKL (1:100; 141607, BioLegend). The stained samples were examined by flow cytometry (FACS LSRFortessa, BD). The data were collected using BD FACSDiva software and analysed by FlowJo (v10.8).
OT-I assay
BMDCs were first pulsed by either 50 nM peptides with or without diABZI or various conjugates for 8 h, or their LNP-encapsulated counterparts for 4 h, and then washed thoroughly before co-culturing with OT-I cells. CD8+ T cells were isolated from OT-I mice by the EasyStep Mouse CD8+ T Cells isolation Kit (19853A, StemCell), stained with CFSE Cell Division Tracker Kit (1:1,000; 423801, BioLegend), and incubated with BMDCs at a 20:1 ratio for 72 h. The data were collected using BD FACSDiva software and analysed by FlowJo (v10.8).
STING–APEX2 assay
HEK293T cells were first treated with the 10 μM cGAS inhibitor G140 1 h before transfection, and then transfected with pcDNA3.1-STING-APEX232. Twenty-four hours later, cells were stimulated with 0.5 μM diABZI, OVA or ABM5–OVA for 2 h. Before harvesting, cells were incubated with 500 μM biotin-phenol (HY-125658, MCE) in a complete medium for 30 min at 37 °C, followed by treatment with 1 mM H2O2 (AAPR43-100, Pythonbio) at room temperature for 2 min. Cells were then washed three times with fresh quencher solution containing 10 mM sodium ascorbate (V900326, Sigma-Aldrich) and 5 mM Trolox (HY-101445, MCE) in PBS. After washing, the cells were lysed in 600 μl of fresh RIPA lysis buffer (HY-K1001, MCE) supplemented with 1 mM PMSF (ST506, Beyotime), 5 mM Trolox and 10 mM sodium ascorbate, assisted by sonification. The lysate was then centrifuged at 13,400g for 20 min at 4 °C to pellet the cell debris. Protein concentration was measured using the BCA assay (P0012S, Beyotime). Clarified cell lysates were incubated with 50 μl of streptavidin magnetic beads (11641778001, Roche) at 4 °C overnight on a rotator to pull down biotinylated proteins. The beads were then washed twice with RIPA lysis buffer, once with 0.2 M KCl, once with 0.1 M Na2CO3, once with 2 M urea in 10 mM Tris-HCl (pH 8.0), and twice again with RIPA lysis buffer at 4 °C. Biotinylated proteins were eluted by boiling the beads in 75 μl of protein loading buffer supplemented with 0.67 mM biotin and 6.7 mM dithiothreitol for 10 min. The eluates were collected by pelleting the beads using DynaMag magnets (12321D, Invitrogen) and analysed by western blot.
Western blot
BMDCs receiving various treatments were lysed by RIPA. BMDC lysate in RIPA or final eluates from the STING–APEX2 assay were prepared in protein loading buffer and stored at −20 °C until use. The membranes were first stained by anti-STING (1:2,000; 13647, CST), anti-TBK1 (1:2,000; 3504T, CST), anti-pSTING (1:500; 72971S, CST), anti-pTBK1 (1:500; 5483T, CST), anti-TAP1 (1:1,000; 11114-1-AP, Proteintech), anti-Proteasome 20S (1:1,000; ab22673, Abcam) and anti-GAPDH (1:3,000; ABL1020, Abbkine). The corresponding horseradish peroxidase (HRP)-conjugated anti-mouse IgG (1:5,000; 7076, CST) or HRP-conjugated anti-rabbit IgG (1:5,000; 7074, CST) were used as secondary antibodies. After extensive washing, the bands were developed using an enhanced chemiluminescent detection kit (180-5001, Tanon) and visualized by an imaging system (ImageQuant 800, Cytiva).
ER isolation and immunodot blot
HeLa cells were transfected by pCMV-STING1(human)-3×FLAG-WPRE-Neo for 24 h, and pulsed by 0.5 μM of OVA–biotin, diABZI + OVA–biotin, ABD–S–OVA–biotin or ABM5–OVA–biotin for 2 h. ER was isolated by an ER isolation kit (ER0100, Sigma-Aldrich). The ER-containing fraction was collected and spotted on the polyvinylidene fluoride (PVDF) membrane. The film was blocked, washed and stained by streptavidin–HRP (1:1,000; 21130, Thermo Scientific), or anti-calreticulin (1:1,000; ab2907, Abcam), followed by washing and staining with HRP-conjugated anti-rabbit IgG (1:5,000; 7074, CST) as secondary antibodies. After extensive washing, the dots were developed using an enhanced chemiluminescent detection kit, visualized by an imaging system.
LNPs
LNPs were prepared by microfluidic mixing. In brief, all lipid materials were dissolved in ethanol at 10 mg ml−1 as stock solutions. The mass ratio of LNP was ionizable lipid SM102 (SN-M-CL23, SUNA Med-Engineering) or ALC0315 (SN-M-CL19, SUNA Med-Engineering):DSPC (LP-R4-076-1, Ruixibio):cholesterol (C3045, Sigma-Aldrich):DSPE-PEG2000 (880120P, Avanti Polar Lipids) at 10:50:37.5:2.5. Peptides, diABZI or ABM5–peptide conjugations were diluted in PBS at 333 μg ml−1 as aqueous phase. The mixture of lipids was used as the ethanol phase. For DiD-labelled LNPs, DiD was dissolved in the ethanol phase at the final concentration of 100 μg ml−1. The ethanol phase and aqueous phase were placed in a LNP packaging system (INano L, Micro & Nano) with a 1-ml or 2.5-ml syringe, squeezed at a total flow rate of 12 ml min−1 with the volume ratio ethanol:aqueous = 1:3. Fresh nanoparticles were then dialysed against PBS through 50 kDa MWCO dialysis bags (AFH0318, Milone) with gentle agitation at 4 °C for 6 h, changing the PBS every 2 h. The size of the LNPs was measured by Particle Analyzer (Litesizer 500, Anton Paar). Encapsulation efficiency was determined by a BCA Protein Assay Kit of LNP-containing solely peptides and compared with the standard curve of peptides under the same conditions. LNPs containing diABZI and SABER–peptide conjugates were quantified by HPLC. The resulting LNPs were stored at 4 °C.
Preparation of ISCOMs
The ISCOMs adjuvant used in this study is a self-assembled, saponin-based nanoparticle prepared as previously described53. In brief, cholesterol and phosphatidylcholine were dissolved in ethanol at the final concentration of 10 mg ml−1, respectively. Meanwhile, 4 mg Quil-A (vac-quil, InvivoGen) was dissolved in 3 ml of sterile PBS. Then, 80 μl of each cholesterol and phosphatidylcholine solution was mixed first, then injected into the Quil-A solution dropwise under the stirring condition. The mixture was allowed to stir for a further 2 h at room temperature, followed by dialysis against PBS using a 10 kDa MWCO dialysis bag (AFH0272, Milone) for 24 h at room temperature. After dialysis, the mixture was further purified by passing through the PD-10 desalting column (Sephadex G-25M, 17085101, GE). The pass-through solution was collected and determined the particle size by Particle Analyzer. Only the fraction with an average particle size of less than 50 nm was collected and used as the adjuvant. The ISCOMs adjuvant was quantified according to the content of Quil-A by HPLC.
Immunization and immune responses
C57BL/6 mice, 6–8 weeks old, were subcutaneously immunized with various free or LNP-encapsulated peptides, with or without free or LNP-encapsulated diABZI, free poly-I:C (tlrl-pic, InvivoGen), ODN1018 (HY-150724C, MCE) or ISCOMs, serving as controls or benchmarks. Peptides and diABZI were used at 10 nmol per mouse. Poly-I:C, ODN1018 and ISCOMs were used at 10 μg per mouse. Other mice were subcutaneously immunized with free or LNP-encapsulated SABER–peptide conjugates at 10 nmol per mouse. All mice were immunized on days 0 and 14, or received a third dose on day 28. Batf3−/− and STING−/− mice were similarly immunized, and WT C57BL/6 mice with the same age and gender were used as controls. The peripheral blood and spleens were collected 7 days after the last injection. The peripheral blood was lysed with ACK lysis buffer twice, and washed with PBS containing 1% FBS. Spleens were processed into single-cell suspensions by passing the tissues through 40-μm cell strainers, lysed with ACK lysis buffer, and the remaining cells were washed with PBS containing 1% FBS. To analyse the CD8+ T cell response, PBMCs and splenocytes were blocked by anti-CD16/CD32 (1:400; 101301, BioLegend), stained with Live/Dead dye (1:400; 423101 or 423113, BioLegend), FITC–anti-CD3 (1:100; 100203, BioLegend), APC–anti-CD8a (1:100; 100712, BioLegend) antibodies, and PE-conjugated OVA-tetramer or Adpgk-tetramer (1:200; HG08T14028, Helixgen). All stained cells were examined by flow cytometry. The data were collected using BD FACSDiva software and analysed by FlowJo (v10.8).
Tumour models
For therapeutic vaccine experiments, all peptides, diABZI and SABER–peptide conjugates were encapsulated in LNPs and used at 10 nmol per mouse, except for poly-I:C, which was used as carrier-free at 10 μg per mouse. B16-OVA, E.G7-OVA or MC38 tumour cells were subcutaneously inoculated at 2 × 105, 5 × 105 and 1 × 105 cells per mouse, respectively. The therapeutic vaccines were subcutaneously injected at distant sites 4, 11 and 18 days after tumour inoculation. In the B16F10 therapeutic vaccine model, tumour cells were subcutaneously inoculated at 1.5 × 105 cells per mouse. Peptides with various adjuvants or SABER–peptide conjugates were subcutaneously injected at distant sites 6, 13 and 20 days after tumour inoculation, in the presence or absence of 200 μg anti-PD1 (BE0146, BioXcell) injected intraperitoneally 1 and 4 days after each immunization. For the B16-OVA prophylactic vaccine model, 2 × 105 tumour cells were subcutaneously injected 7 days after the last immunization. Tumours were monitored by digital callipers, and the tumour volumes were calculated as 1/2 × L × W2, in which L and W are the long and short diameters of the tumours, respectively. When the tumour size exceeded 1,500 mm3 or the longest diameter exceeded 1.5 cm, the last data were recorded and mice were then euthanized. Tumour sizes were maintained in compliance with Institutional Animal Care and Use Committee guidelines (up to 2 cm in one dimension) in any of the experiments. For the B16-OVA lung metastasis model, mice were injected intravenously with 2 × 105 tumour cells 14 days after the last immunization. Mice were euthanized 18 days after tumour injection, and metastatic foci of the lungs were counted. For MC38 tumour rechallenge, 1 × 105 MC38 cells were subcutaneously injected at distant sites in mice with complete tumour regression 90 days after the primary tumour inoculation. Naive mice without previous tumour inoculation were challenged with 1 × 105 MC38 cells as controls. To analyse MC38-specific memory T cells, mice were euthanized 60 days after rechallenge. Splenocytes were blocked by anti-CD16/CD32 (1:400; 101301, BioLegend), stained with Live/Dead dye (1:400; 423107, BioLegend), FITC–anti-CD3 (1:100; 100203, BioLegend), APC–anti-CD8a (1:100; 100712, BioLegend), PB–anti-CD44 (1:100; 156005, BioLegend), APC/Cy7–anti-CD62L (1:100; 104427, BioLegend), BV650–anti-CD127 (1:100; 135043, BioLegend) and PE-conjugated Adpgk-tetramer (1:200; HG08T23002, Helixgen). All stained cells were examined by flow cytometry. The data were collected using BD FACSDiva software and analysed by FlowJo (v10.8).
Biodistribution of LNPs
B16-OVA tumour cells were subcutaneously inoculated into C57BL/6 mice. When tumour size achieved 200 mm3, DiD-labelled LNP-encapsulated AMB5–OVA was subcutaneously injected at 10 nmol per mouse. The mice were euthanized 24 h later. The heart, liver, spleen, lungs, kidneys, drain lymph node and tumour were collected and imaged by an imaging system (IVIS Lumina III, PerkinElmer). To identify DC subtypes taking DiD-labelled LNPs, draining lymph nodes were collected 24 h later. Single cells were obtained by passing the tissues through 40-μm cell strainers, lysed with ACK lysis buffer, blocked by anti-CD16/CD32 (1:400; 101301, BioLegend), and stained with FITC–anti-CD11c (1:100; 117306, BioLegend), BV711–anti-CD8a (1:100; 100747, BioLegend), APC/Cy7–anti-CD103 (1:100; 121431, BioLegend), PE–anti-CD11b (1:100; 101207, BioLegend), PE/Cy7–anti-H2-Kb(SIINFEKL) (1:100; 141607, BioLegend) and PB–anti-CD86 (1:100; 105021, BioLegend). The data were collected using BD FACSDiva software and analysed by FlowJo (v10.8).
Injection site local reactions, blood chemistry and cytokines
C57BL/6 mice were subcutaneously immunized with 10 nmol of LNP-encapsulated ABM5–OVA or diABZI + OVA. The injection sites were photographed at the indicated time. Serum was collected on days 0, 1, 4 and 8. Mouse IFNβ and TNF were measured by ELISA kits (439404 and 430904, BioLegend). Blood chemistry, including alanine transaminase (ALT), aspartate transferase (AST), creatinine and urea levels, was examined on days 0, 8 and 14 by an auto-chemistry analyzer and associated kits (BS-240VET, Mindray). Normal ranges of blood chemistry in C57BL/6 mice were delineated according to the manufacturer’s manual in the kit.
SARS-CoV-2 vaccination and challenge
In the experiments using ABM5–SNT as a standalone vaccine, C57BL/6 or hACE2-Tg mice were subcutaneously immunized with 30 nmol of LNP-encapsulated ABM5–SNT or diABZI + SNT on days 0 and 14. In the experiments using ABM5–SNT as an adjuvant for subunit vaccines, C57BL/6 mice were subcutaneously immunized with 10 μg RBD-Fc or 100 μg OVA, in the presence of 10 nmol LNP-encapsulated ABM5–SNT or diABZI + SNT on days 0 and 14. Spleens and sera were collected on day 21. The intracellular cytokine assay for SARS-CoV-2 N protein antigens was performed according to a previous study54. In brief, splenocytes were stimulated with 8 μg ml−1 SNT or 2 μg ml−1 SARS-CoV-2 spike peptide pool (PP003, Sino Biological) for 6 h in the presence of 4 μg ml−1 anti-CD28 (1:400; 102102, BioLegend) and Cell Stimulation Cocktail (1:500; 00-4970-93, Invitrogen). Brefeldin A (10 μg ml−1; S1536, Beyotime) was added to the cell culture 4 h before harvest. Cells treated with anti-CD28, cocktail and brefeldin A but without peptide stimulation served as a negative control. Cells were stained with Live/Dead dye (1:400; 423101, BioLegend), blocked by anti-CD16/CD32 (1:400; 101301, BioLegend) and stained with antibodies against T cell markers. Then, cells were fixed and permeabilized by Fix/Perm solution (554714, BD), washed with Perm/Wash buffer (554723, BD) and stained by antibodies to cytokines. For CD8+ T cells, FITC–anti-CD3 (1:100; 100203, BioLegend), PE–anti-CD8b.2 (1:100; 140408, BioLegend) and APC–anti-IFNγ (1:100; 505810, BioLegend) were used. For CD4+ T cells, FITC–anti-CD3 (1:100; 100203, BioLegend), PE–anti-CD4 (1:100; 100408, BioLegend), APC–anti-IFNγ (1:100; 505810, BioLegend) and PE/Cy7–anti-IL-4 (1:100; 504117, BioLegend) were used. All stained cells were examined by flow cytometry. The data were collected using BD FACSDiva software and analysed by FlowJo (v10.8).
The SARS-CoV-2 challenge experiment was conducted as previously reported38,55. The vaccinated hACE2-Tg mice were anaesthetized and challenged with intranasal instillation of 5,000 plaque-forming unit SARS-CoV-2 Omicron BA.5.2 variant. The behaviour and clinical signs of the mice were recorded daily after infection. Mice were euthanized to dissect lung and brain tissues for viral load determination 4 days after the challenge. Tissues were homogenized in TRIzol (15596026, Thermo Fisher) using an electric homogenizer. Total RNA was extracted and the SARS-CoV-2 subgenomic E gene was quantified by RT–qPCR using specific primers and probes: SGMRNA-E-F: CGATCTCTTGTAGATCTGTTCTC; SGMRNA-E-R: ATATTGCAGCAGTACGCACACA; and SGMRNA-E-probe: FAM-ACACTAGCCATCCTTACTGC GCTTCG-BHQ1. The SARS-CoV-2 E gene was cloned into the pcDNA3.1 vector and a standard plasmid was constructed. The viral load in the samples was determined and calculated from the standard curves.
ELISpot
Splenocytes were stimulated with 8 μg ml−1 SNT peptides. Mouse IFNγ precoated ELISpot kit (DKW22-2000-096, Dakewe) was used for the analysis of IFNγ production on hydrophobic PVDF. Spots were counted by an ELISpot reader (S6 Ultra, C.T.L.).
ELISA
An indirect ELISA was performed to determine antibody titres against OVA or WT RBD. A 96-well plate was coated with 1 μg of OVA or 0.1 μg RBD protein per well, blocked with 5% BSA and washed by PBST (0.05% Tween-20 in PBS). Mouse serum was diluted in PBST and incubated with the coated plates. After washing, HRP-conjugated anti-mouse IgG (1:3,000; 7076, Cell Signaling Technology) was incubated as secondary antibodies. After extensive washing, antigen-specific IgGs were quantified by using 3,3′,5,5′-tetramethylbenzidine (P0209, Beyotime) and measuring OD450 by a microplate reader (Varioskan Lux, Thermo Fisher). End point titres were determined as the highest reciprocal serum dilution with optical density at 450 nm (OD450) value 2.1-fold over the background.
Pseudovirus neutralization assay
Generation of pseudoviruses and the neutralization assay were performed as previously described56,57. In brief, pseudoviruses expressing S protein of WT, Delta, BA.1 or BA.5 strains were obtained by transfection of pcDNA3.1-spike (WT, Delta, BA.1 or BA.5), psPAX2 and pLenti-CMV-Puro-Luc (168w-1) plasmids into HEK293T. The culture supernatants containing luciferase-expressing pseudoviruses were harvested, titrated and stored at −80 °C until use. The hACE2-293T cells (2 × 104 cells per well) were seeded in the black flat-bottom 96-well plates. Heat-inactivated sera were first diluted 25-fold then 4-fold serially diluted in DMEM, and subsequently co-incubated with the same volume of pseudoviruses for 1 h at 37 °C. The mixtures were supplied with 10 μg ml−1 polybrene (C0351, Beyotime) to hACE2-293T cells for 6-h absorption. The mixtures were discarded, and fresh culture medium was added and incubated for an additional 42 h. After incubation, the infected cells were lysed using firefly luciferase lysis buffer (RG126M, Beyotime) and incubated with the luciferase substrate (RG058M, Beyotime). The relative light unit was measured using a microplate reader. The half pseudovirus neutralization titres were determined with a four-parameter non-linear regression curve by GraphPad Prism (v9.5.1).
Statistical analysis
All statistical analyses were performed using GraphPad Prism (v9.5.1). Data were shown as mean ± s.e.m., geometric mean ± 95% CI, violin plot, or box and whiskers as indicated in each experiment. Comparisons of two groups were assessed using two-tailed Student’s t-test. Comparisons of multiple groups were assessed using one-way ANOVA with Tukey’s multiple comparisons test, Kruskal–Wallis with Dunn’s multiple comparisons test or two-way ANOVA. Survival curves were compared using the log-rank test. The investigators were not blinded to the experiments that were carried out under highly standardized and predefined conditions, except for microscopy images and tumour measurement, which were evaluated in an investigator-blind manner.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.