Inclusion and ethics
This study was approved by the Ethics Committee of Westlake University (approval no. 20200722YJ001).
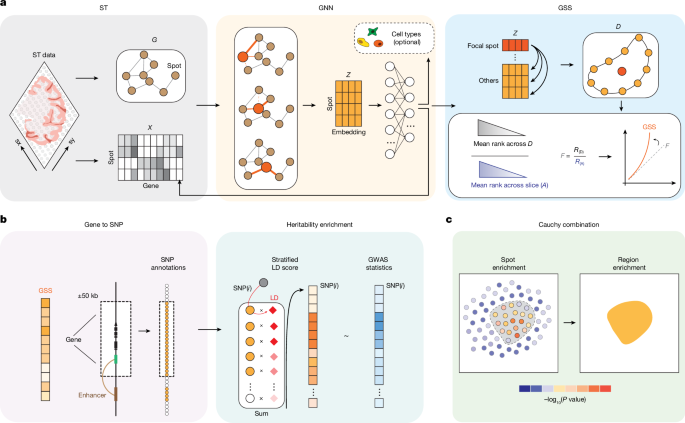
gsMap method
Design principles
To effectively illustrate gsMap, we first summarize its design principles. gsMap utilizes the framework of S-LDSC17 to assess whether genetic variants, mainly SNPs, located in or near genes specifically expressed in a spot in ST data are enriched for genetic associations with a trait of interest. To precisely estimate gene expression specificity for individual spots, gsMap aggregates information from homogeneous spots, a crucial step given the sparsity and high technical noise in gene expression profiles of individual spots in ST data12,55. Using spatial coordinates alone is inadequate to identify homogeneous spots because spatially neighbouring spots may not necessarily belong to the same cell type. Using gene expression profiles alone could also lead to biased identification of homogeneous spots owing to technical noise. To address these limitations, gsMap uses a GNN to learn embeddings that integrate spatial coordinates and gene expression profiles, and then identifies homogeneous spots for each focal spot on the basis of their similarity in the embedding matrix. gsMap then estimates GSSs for each focal spot by aggregating information from its homogeneous spots. Note that although spot embeddings for ST data generated from existing tools56,57 can be applied in gsMap, to enhance usability of our software tool and improve embedding quality, we developed a built-in GNN model (see below) that incorporates spot annotations when available and uses a graph attention layer (GAT) to prevent over-smoothing during the process of identifying homogenous spots for a focal spot.
Data input
gsMap requires inputs of (1) GWAS summary statistics; (2) sequencing-based ST data comprising transcriptome-wide gene expression profiles and spatial coordinates of individual spots; (3) LD reference data; and (4) optionally, SNP-to-gene linking maps. We noted that the resolution of ST data varies significantly across different platforms, ranging from one spot on sequencing array chips representing a cluster of cells (for example, 10X Visium) to spots at sub-cellular resolution (for example, Stereo-seq). To ensure the robustness of gsMap when handling ST data at sub-cellular resolution, cell segmentation58 analysis is required, which merges original spots on sequencing array chips into individual cells. For simplicity, we continue to use spots to denote data points in ST data, where one spot represents an individual cell (after cell segmentation analysis) in high-resolution ST platforms or multiple cells in conventional ST platforms.
Processing gene expression data
The gene expression count matrix is log-transformed and normalized according to the library size of each spot. Subsequently, the normalized gene expression matrix is standardized to attain a zero mean and unit variance, and the top h (default, 3,000) highly variable genes (HVGs) are retained. We select HVGs based on the normalized variance of each gene, which adjusts for mean-variance associations, as implemented in Scanpy59. The resulting processed gene expression matrix is represented as \(X\in \mathbbR^n\times h\), with n denoting the number of spots and h representing the number of HVGs. To accelerate the training process in the GNN, users can optionally provide a principal component analysis-reduced matrix (default, 300 components). The normalized HVG matrix (or principal component analysis-reduced matrix) serves as the feature matrix in the GNN.
Building a spatial graph of the spots
The spatial coordinates of individual spots are transformed into an undirected graph, denoted as \(G=(V,E)\). In this graph, each vertex \(v\in V\) represents a spot, and E represents the set of connected edges between spots. For balancing the performance and computational efficiency, gsMap, by default, considers the ten nearest neighbouring spots for each spot, determined by their Euclidean distance in spatial coordinates. The resulting graph can be delineated by the adjacency matrix \(A\in \mathbbR^n\times n\) with n denoting the number of spots. If spots i and j are connected in the graph, \(A_ij=1\); otherwise, \(A_ij=0\).
Learning embedding matrix
The standardized gene expression matrix X and the adjacency matrix A are then integrated into an embedding matrix \(Z\in \mathbbR^n\times m\), where m represents the number of features (set as 32), using the GAT auto-encoder framework. The advantage of GAT60 lies in its trainable edge weights among connecting spots, enabling higher weights for spots with analogous gene expression patterns during information aggregation. A detailed description of the graph attention auto-encoder can be found in section 1 of the Supplementary Note.
The loss function of the graph attention auto-encoder in gsMap includes two parts: (1) the mean squared error used for reconstructing gene expression matrix; and (2) the cross-entropy loss used for predicting cell types of spots:
$$L=\gamma \left(\mathop\sum \limits_i=1^n\mathop\sum \limits_g=1^h(x_ig-\widehatx_ig)^2\right)+(1-\gamma )\left(\mathop\sum \limits_i=1^n\mathop\sum \limits_k=1^c-p_ik(q_ik)\right)$$
Here, \(x_ig\) represents the normalized expression value of spot i for gene g with \(\widehatx_ig\) being the reconstructed value. \(p_ik\) represents the probability of spot i belonging to cell type k, coded as 1 if spot i is of cell type k, and 0 otherwise. \(q_ik\) represents the predicted probability, c is the number of cell types, and γ is the hyperparameter that balances the reconstruction loss and cross-entropy loss. In practice, γ is set to 0.5 in most cases. In scenarios where annotated cell types are unavailable, γ is set to 1. Of note, cell type annotations are included to improve embedding accuracy, but they are not strictly necessary, as they can largely be captured by gene expression profiles (Supplementary Fig. 22). During the training process, the Adam optimizer61 is utilized to minimize the loss function and the exponential linear unit (ELU)62 is used as the activation function. The weight decay is set to 10−4, and the maximum number of iterations is set to 1,000. The iteration is considered as converged when \(| L^t+1-L^t| < 10^-4\).
Identification of homogeneous spots
The embedding matrix Z integrates information from gene expression values, spatial locations of spots, and cell type priors. We then identify homogeneous spots for each focal spot on the basis of their cosine similarity in this latent space:
$$\cos (\theta _ij)=\frac\bfZ_i^\rm\top \bfZ_j\parallel \bfZ_i\parallel \parallel \bfZ_j\parallel $$
where \(\bfZ_i\in \mathbbR^m\times 1\) is the embedding vector of spot i. For each focal spot, we select the top d spots that show the highest cosine similarity with it. This process aligns each individual spot with d spots that are spatially close and share similar transcriptomic profiles, referred to as the microdomain (D). In this study, we set d to 20 for ST data generated from the 10X Visium platform and 50 for ST data generated from the Stereo-seq platform.
Estimation of GSS
We rank genes in individual spots on the basis of their expression levels, with higher expression values receiving higher ranks. We opt to use gene expression ranks instead of gene expression values because the ranks are more robust against artefacts across different technical and biological replicates (Supplementary Fig. 23), as previously suggested63. For each gene, its expression specificity within each focal (individual) spot is assessed by calculating the geometric mean of its expression rank across the microdomain of the focal spot, divided by the geometric mean of its expression rank across all spots in the ST data. For gene g, \(F_ig\) represents its expression specificity in spot i, calculated as:
$$F_ig=\frac\sqrt[d+1]R_ig(\,\prod _k\in D_iR_kg)\sqrt[n]\mathop\prod \limits_j=1^nR_jg$$
where \(R_ig\) denotes the expression rank of gene g in spot i, and Di represents the microdomain (that is, set of homogonous spots) of spot i with d denoting the spot number. If \(F_ig < 0\), indicating that this gene is not specifically expressed within the focal spot, we set it to 0. Additionally, we compare the expression proportion of each gene across the focal spot microdomain to its proportion across all spots. If this ratio is smaller than 1, suggesting that the large \(F_ig\) might be due to outliers with discordant high expression ranks, we also set \(F_ig\) to 0. To align the scale between GWAS summary statistics and the estimated gene specificity, we performed an exponential projection to further distinguish genes with high expression specificity:
$$S_ig=\exp \F_ig^2\-1$$
where \(S_ig\) denotes the final specificity score of gene g in spot i.
Mapping GSSs to SNPs
We map the specificity score of each gene to the corresponding SNPs within a window extending 50 kb upstream and 50 kb downstream of each gene’s transcribed region. We have shown through a sensitive analysis that gsMap is robust to different window sizes (Supplementary Fig. 24). We have included an option in the gsMap software tool for mapping SNPs outside the 100-kb window to genes, based on SNP-to-gene linking maps derived from epigenomic data (for example, Roadmap15 and Activity-by-Contact model16). This process yields a unique set of SNP annotations for each spot. On average, across the ST datasets used in this study, there are more than 150,000 SNPs and 3,000 genes with a non-zero GSS per spot.
Linking genomic annotations with GWAS data
Treating each spot as a set of SNP annotations, gsMap assesses whether SNPs with higher GSS are enriched for heritability for the trait of interest using the S-LDSC17 framework, conditional on the baseline SNP annotations. The S-LDSC in gsMap can be considered as a linear regression analysis between GWAS χ2 statistics and stratified LD scores computed using SNP annotations from individual spots. A detailed explanation of S-LDSC can be found in section 2 of the Supplementary Note. The LD reference data used in this study were obtained from the 1000 Genomes Project Phase 3 (1KGP3)64. Following previous studies10,65, we use block-jackknife to estimate the standard error of the regression coefficient in S-LDSC. P value is computed using a one-sided z-test, assessing whether the regression coefficient is significantly larger than 0. A smaller P value indicates a stronger relevance of the focal spot to the trait of interest.
Estimating the strength of enrichment for a spatial region
To evaluate relevance between a specific spatial region and the trait of interest, gsMap uses the Cauchy combination test19 to aggregate P values of individual spots within the spatial region:
$$T_\rmCauchy=\mathop\sum \limits_i=1^\phi \tan \(0.5-P_i)\rm\pi \$$
where \(P_i\) represents the P value of spot i belonging to the spatial region. The aggregated P value for the spatial region is approximated as:
$$P_\rmregion\approx \frac12-\rm\pi \\arctan (T_\rmCauchy)\$$
The Cauchy combination test combines signals from all spots within a spatial domain, demonstrating high consistency (median r = 0.82) with the previous domain-level approach13 while offering improved statistical power (Supplementary Fig. 21).
Running time
We have optimized the gsMap code to ensure its efficiency in handling large ST data. gsMap analyses large ST datasets with 120 K spots within 3 CPU hours, whereas even with the single-spot GSS available, using S-LDSC software for the same task would require over 20,000 CPU hours (~10 min per spot). A summary of the gsMap runtime for each step is available in section 3 of the Supplementary Note.
Diagnostic tools
We have included a module in the gsMap software tool, which enables users to plot spatial distributions of expression levels and GSS for selected genes, as well as generate Manhattan plots highlighting SNP association signals mapped to these genes, aiding in the diagnostic analysis of the data (Supplementary Fig. 25).
Joint analysis mode
We have developed a joint analysis mode to analyse ST data with multiple technical and/or biological replicates, producing unified results and simplifying interpretation (Supplementary Fig. 26). In brief, a GSS is calculated for each spot as the mean rank of a gene within homogeneous spots, normalized by its mean rank across all spots (including those from different biological replicates). This step harmonizes GSS annotations across biological replicates. We then perform heritability enrichment analysis (for example, using S-LDSC) to associate each spot with traits and apply the Cauchy combination test to combine P values across all spots with the same annotations
Quantifying the association of a spatial region with a trait relative to all other regions
We used an OR, computed as the ratio of trait-associated spots to non-associated ones in a specific region divided by the ratio in all other regions, to compare gsMap results of a specific spatial region across traits with varied GWAS statistical power. This metric quantifies the strength of a region’s association with a trait, relative to all other regions. Consequently, it compensates for differences in gsMap results owing to variations in GWAS statistical power, allowing for a more meaningful comparison of gsMap results across traits. The significance of the OR was evaluated using a Chi-square test.
Simulations
The simulation study was conducted using real genotype data on 100,000 unrelated individuals of European ancestry from the UK Biobank22. We used the HapMap3 SNPs and filtered out SNPs with a minor allele frequency (MAF) < 0.01 or Hardy–Weinberg equilibrium test P value < 10−6, resulting in a total of 1,195,548 SNPs. We used GCTA (V1.94.1)66 to generate quantitative traits based on real genotype data of a set of selected causal variants. We simulated four null scenarios where causal variants were: (1) randomly distributed across the genome; (2) enriched in the candidate cis-regulatory elements (cCREs)67; (3) enriched within LD blocks; and (4) enriched in the cis-regions of non-spatially variable genes (NSVGs), respectively. Here ‘null’ means that the simulated causal variants are not enriched in or around genes with context-dependent expression, meaning that the simulated trait is not expected to be associated with any specific group of spots.
In scenario 1, we simulated phenotypes with varying levels of polygenicity (that is, proportion of SNPs being causal) and heritability (that is, proportion of variance in the phenotype attributed to the causal SNPs). The number of causal SNPs varied from 10,000 to 500,000, and the heritability ranged from 0.1 to 0.6. In scenarios 2 to 4, we fixed the number of causal SNPs at 100,000 and heritability at 0.3. For scenario 4, we used two strategies to select NSVGs. First, we chose genes with a maximum GSS smaller than 1.5, computed by gsMap. Second, to select genes entirely independent of the gsMap analysis process, we used SPARK-X68 to test for spatially dependent expression and selected genes with a P value greater than 0.85. Each simulation was replicated three times. We used PLINK (V1.90)69 to associate SNPs with the simulated phenotypes with the first ten principal components, derived from SNPs, fitted as covariates.
GWAS summary statistics
We collected GWAS summary statistics from the public domain for a broad range of complex traits and diseases, spanning eight major categories: autoimmune, psychiatric, reproductive, behavioural, metabolic, haematological, anthropometric and cancer. To ensure sufficient GWAS power, we included only those traits for which the Chi-square statistic for the LDSC estimate of heritability exceeded 25, as suggested by prior work8,10. In summary, we analysed GWAS summary statistics for 110 complex traits, including diseases, from the UK Biobank and other publicly available sources (average n = 385,000, Supplementary Table 1). We excluded the major histocompatibility complex (MHC) region from all analyses owing to its complexity17.
Spatial transcriptomics datasets
We included six spatial transcriptomics datasets in this study: mouse embryonic data (Stereo-seq)5, human embryonic data (Stereo-seq)7, mouse brain data (Stereo-seq and MERFISH)5,27, macaque cortical data (Stereo-seq) and human DLPFC data (10X Visium)13. To align mouse or macaque genes with human genes, we utilized the biomaRt (V3.18) R package to conduct homologous gene transformations. The average gene numbers after homologous transformation are 16,330 for the mouse datasets and 13,536 for the macaque dataset. Following the standard analysis pipeline, we utilized the Scanpy (V1.9.6)59 Python package to process each ST dataset. Details of each dataset are summarized below.
Mouse embryonic ST data
We analysed 54 coronal sections from the mouse embryonic dataset, sourced from eight C57BL/6J mice, with an average of 81,125 spots per section, spanning from the embryonic stage of E9.5 to E16.55. Data at both bin50 resolution (53 sections) and single-cell resolution (1 section) were included in this study. We obtained access to the h5ad files, each including the gene expression count matrix, cell type annotations, and spot spatial coordinates for a section. We validated the cell type annotations based on known marker genes.
Human embryonic ST data
We analysed 62 transverse sections from the human embryonic dataset, sourced from a CS8 human embryo of Chinese ancestry, with a total of 38,562 spots7. We had access to the h5ad files, each containing the gene expression count matrix, cell type annotations, and spot spatial coordinates.
Mouse brain ST data
We analysed two sections from the mouse brain dataset: one coronal section from an adult C57BL/6J mouse brain (50,140 spots) and one sagittal section from an E16.5 embryonic C57BL/6J mouse brain (65,303 spots)5. Both sections are at single-cell resolution. We had access to the h5ad files and verified the cell type annotations using known marker genes.
Mouse brain MERFISH ST data
We analysed a mouse brain MERFISH ST dataset27 consisting of two sections: a coronal section from an adult C57BL/6J-1 mouse brain containing 41,181 spots and a sagittal section from an adult C57BL/6J-3 mouse brain containing 105,934 spots. This dataset was generated using an image-based platform, where a panel of ~1,100 genes was imaged. The remaining genes were imputed by the authors using paired scRNA-seq data. We accessed the imputed ST dataset, which includes expression levels for 15,768 genes, cell spatial coordinates, cell type annotations, and brain region annotations.
Macaque cortical ST data
We analysed 162 coronal sections from the macaque cerebral cortical dataset, sourced from three adult male cynomolgus monkeys, with an average of 266,654 spots per section35. All sections are at single-cell resolution. We obtained access to the sctransform70 gene expression matrices and the metadata files. For each section, we aligned the spots spatial coordinates, cell type annotations, cortical region annotations, and section-cutting positions (EBZ) and compiled all the aforementioned information into an h5ad file.
Human DLPFC ST data
We analysed eight coronal sections from the adult human DLPFC dataset, sourced from two donors of European ancestry, with an average of 3,973 spots per section13. These data were generated using 10X Visium, with each spot containing a few dozen cells. We had access to the h5ad files, each including the gene expression count matrix, spots spatial coordinates, and cortex layer annotations.
Single-cell RNA-seq datasets
We used six scRNA-seq datasets in this study to estimate gene expression correlations between mice and humans47,71,72,73,74,75. These datasets contain 9 million cells, covering 25 major tissues from both mice and humans. The cell type annotations were provided by the authors of the scRNA-seq datasets, and we re-clustered them into eight primary cell categories: endothelial cells, epithelial cells, glial cells, neurons, immune cells, muscle cells, stem cells and others.
Comparison of the gsMap results from the human and macaque datasets
We applied gsMap to eight human DPLFC ST sections and observed highly consistent results across these sections. We then calculated an OR for each cortex layer, representing the strength of association of a cortex layer with a trait, relative to all other layers. To ensure a robust comparison between the human and macaque results, we compared the median OR value of each cortex layer across eight human DPLFC ST sections to that across nine macaque PFC ST sections. Considering that there were only five matched cortex layers between the human and macaque datasets, we pooled the OR values from 22 brain-related traits, resulting in 110 data points in the comparison analysis.
Integration of GWAS summary statistics with ST data using scDRS
scDRS9 is a method that can integrate GWAS summary statistics with scRNA-seq data to identify cells relevant to a trait. In brief, scDRS computes a trait-enrichment score to examine whether a cell has excess expression levels across a set of trait-associated genes. These genes were derived from GWAS summary statistics using gene-based association tests (for example, MAGMA76). To assess the statistical significance of the trait-enrichment score, a P value is calculated by comparing the trait-enrichment score to those computed from control genes with matched expression characteristics. Though originally developed for scRNA-seq data, scDRS can, in principle, be applied to ST data by regarding each spot in the ST data as a cell. Following the standard analysis protocol of scDRS (V1.03), we first used MAGMA to generate gene-based test statistics from the GWAS summary statistics, utilizing reference LD data obtained from 1KGP3, as done in gsMap analysis. Next, we used the ‘munge-gs’ command in scDRS and set the ‘n-max’ parameter to 1,000, to generate a gene-weight file for the top 1000 trait-associated genes identified from the MAGMA analysis. Finally, we used the ‘compute-score’ command in scDRS and set the ‘n-ctrl’ parameter to 1,000, which generated 1,000 sets of control genes using Monte Carlo sampling, to obtain the trait-enrichment P value for each spot.
Expression association analysis
We performed two expression association analyses to identify genes whose expression levels in Glu-neurons were associated with their positions along the CA1 D–V axis, and genes in Glu-neurons whose expression levels were associated with their relevance to SCZ.
Expression association analysis-1 (for gene g):
$$\bfg=\bfx\beta +\bfc\alpha +e$$
where \(\bfg\in \mathbbR^k\times 1\) represents the normalized expression levels of a gene in k Glu-neurons, \(\bfx\in \mathbbR^k\times 1\) represents the spatial positions of k Glu-neurons along the D–V axis, with β being the coefficients, \(\bfc\in \mathbbR^k\times 1\) represents the spot library size vector (a sum of all UMI counts per spot) to adjust for the gene expression variation attributed to spot library size, and e represents the residuals.
Expression association analysis-2 (for gene g):
$$\bfg=\bfz\gamma +\bfc\alpha +e$$
where \(\bfz\in \mathbbR^k\times 1\) represents the −log10 gsMap P values of associations between k Glu-neurons and a trait of interest (for example, SCZ), with γ being the coefficients. The other parameters are defined as above.
Gene prioritization for complex traits
We compared the genes prioritized by gsMap to those identified by other gene prioritization methods, including COLOC77, FUSION78, SMR79 and PoPS80 for the psychiatric disorders (including depression, bipolar disorder and schizophrenia) for which a number of drug target genes are available for validation. We used all eQTL datasets from GTEx81 for FUSION (V1.0.0), all eQTL datasets from GTEx81 and MetaBrain82 for COLOC (V5.2.3) and SMR (V1.3.1), and the feature matrix provided by the PoPS authors for PoPS (V0.2).
Psychiatric drug targets
We collected drug target genes from the DrugBank database43 and Drug Repurposing Hub database42. For the DrugBank database, we selected drugs intended to treat psychiatric disorders categorized under the International Classification of Diseases (ICD) codes F00 to F99. From the Drug Repurposing Hub database, we selected drugs categorized under the psychiatric disease areas. In total, we identified 417 target genes from drugs that are either approved or undergoing clinical trials for treating psychiatric disorders. Based on the identified drug target genes, we used Fisher’s exact test to assess whether genes highly expressed in the macaque PFC 14r region are enriched in these drug target genes, compared to all detected genes, or genes highly expressed in other PFC regions. The module score of these 417 drug targets genes for individual spots was computed using the AddModuleScore function in the Seurat (V4.4.0) R package with the default settings83,84
GO term enrichment
We performed GO term enrichment analysis using the clusterProfiler85 (V3.18) R package with the default settings.
Genetic correlation
We used the bivariate LDSC65,86 (V1.01) to estimate genetic correlations between trait pairs. The reference LD data used in the genetic correlation analysis was generated from 1KGP3.
Statistics and reproducibility
We analysed only existing datasets without using any statistical methods to predefine the sample size. Our study did not involve a design requiring randomization or blinding. We validated our results by performing the same analyses on independent datasets, and all replication analyses were successful.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.