Mice
All animal experiments complied with the regulatory standards of, and were approved by, the University of California Berkeley Institutional Animal Care and Use Committee. Mice were maintained in specific-pathogen-free conditions under a 12 h–12 h light–dark cycle at 20–26 °C and 30–70% humidity, and given water and standard chow diet (Harlan’s irradiated chow) ad libitum. All mice were bred in-house. C57BL/6 and B6.129S2-Ifnar1tm1Agt/Mmjax (Ifnar−/−) mice were originally from Jackson Laboratories and MMRC and further bred in-house. The generation and genotyping of Sp140−/− mice was previously described4. HA-Sp140 knock-in mice were generated by electroporation of C57BL/6 zygotes with Cas9, Alt-R CRISPR-Cas9 crRNA from IDT (gRNA sequence: ACUCCAAGGGACCCUGUUCA), and a homology repair Alt-R HDR donor oligo from IDT (CCCCTGAAGGAGTTCTCTCTGGGCTTCCCAGAGACTCAGAGGGGGTTCGGTCTAGTCTGAACAGGGTCCCTTGGAGTCTGTGTAGGGGATGTACCCATACGATGTTCCAGATTACGCTGCAGGAGGCTACAATGAACTCAGCAGCAGGTAAGTCCCATTCTCTCTTGTCCCTTGTCTC) as previously described61. Founders were backcrossed to C57BL/6J, and mice with matching HA-Sp140 alleles were further bred. HA-Sp140 knock-in mice were genotyped using a qPCR-based assay from Transnetyx. Sp140−/−Resist1−/−Resist2−/− mice were generated by electroporation of Sp140−/− zygotes with Cas9 and sgRNA (CGACGATGGCGGTGACTACC)4. Founders were genotyped and backcrossed to Sp140−/− mice, and progeny with matching alleles were further bred. For genotyping Resist1/2, large ear clips were obtained and digested in QuickExtract lysis buffer overnight with 0.4 mg ml−1 proteinase K, followed by heat inactivation (85 °C for 4 min, 98 °C for 2 min) and stored at −80 °C. After freezing, ear clip lysates were vortexed. Mice were genotyped for Resist1 (Gm21188) with Q5 2× PCR mix according to the manufacturer’s instructions with 1 μl of lysate per 20 μl PCR reaction (F, TTGAGAAATCCGTTTGTAATGGG; R, GCCTTTCTCCGGATTCACGA; cycling conditions: 98 °C for 3 min, 35 cycles of 98 °C for 10 s, 63.5 °C for 20 s and 72 °C for 65 s, followed by a final extension of 72 °C for 10 min). Mice were genotyped for Resist2 (Gm36079) with Phusion GC rich PCR components according to the manufacturer’s instructions, using 1 μl of lysate per 20 μl PCR reaction (F, TGGTATTCTCTAGAGATAACATCACAGCACCTACTTACTCC; R, CCTCCCCTCGCCATCACTGCCTG; cycling conditions: 98 °C for 30 s, 30 cycles of 98 °C for 10 s, 72 °C for 15 s and 72 °C for 60 s, followed by a final extension of 72 °C for 10 min). A total of 5 μl of PCR product was cleaned with FastAP and ExoI, then diluted twofold with water and Sanger sequenced (Resist1 R, GCCTTTCTCCGGATTCACGA; Resist2 F seq, CTGAATGATTCTTCTACTGCTTCCATCC). Sanger sequencing results were evaluated with Snapgene (v.7.0.1).
Cell culture
HEK293T and GP2 cells were obtained from the UC Berkeley Tissue Culture facility and further propagated at 37 °C and 5% CO2 in complete DMEM (10% FBS (v/v) (Gibco) and 1× penicillin–streptomycin and glutamine (Gibco)). BlaER1 cells (from the laboratory of V. Hornung) were cultured in the B cell stage in complete RPMI, and differentiated into monocytes as described previously29. Cell lines tested negative for mycoplasma by PCR (F, CACCATCTGTCACTCTGTTAACC; R, GGAGCAAACAGGATTAGATACCC) with Dreamtaq PCR reagents and validated with short-tandem-repeat profiling by the UC Berkeley Tissue culture facility. HEK293T cells were last validated on 25 November 2024, GP2 cells were last validated on 23 May 2024 and BlaER1 cells were last validated on 23 May 2024.
BMM generation and stimulation
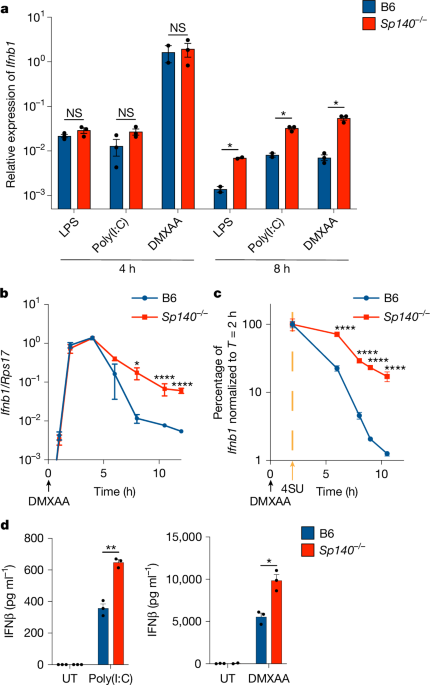
Mice were euthanized and bones (femurs and tibias) were extracted with stringent washing in 70% ethanol. After washing in 70% ethanol and BMM medium (complete RPMI with 10% MCSF (v/v) generated from 3T3 cells as described previously4), bones were crushed in a sterilized mortar and pestle, and bone marrow was passed through a 70-μm filter. Bone marrow from one mouse was divided across eight 15 cm non-tissue-culture-treated plates in 30 ml total volume in BMM medium. The day of bone marrow collection was considered to be day 0, and BMMs were fed on day 3 with 10 ml medium per plate. On day 6, BMMs were collected in cold PBS by scraping and seeded onto non-tissue-culture-treated plates at the appropriate density (100,000 cells per non-tissue-culture-treated 12-well and 24-well; 6-well for over 100,000 cells per well) and rested at least overnight before stimulation. To stimulate, the medium was aspirated and replaced with BMM medium containing 10 ng ml−1 LPS (Invivogen, tlrl-3pelps), 100 μg ml−1 poly(I:C) (Invivogen, tlrl-picw), 100 μg ml−1 DMXAA (Cayman Chemicals, 14617) or 10 ng ml−1 IFNγ (BioLegend, 575304).
RT–qPCR
RNA was isolated using the Omega Biotek Total RNA II kit according to the kit instructions. RNA was subsequently DNase-treated either on-column (Qiagen, 79254) or with RQ1 (Promega, M6101) according to the manufacturer’s instructions. For BlaER1 cells, RNase inhibitor was always included with DNase treatment (either RNaseOUT (Invitrogen) or RNasin (Promega)). RNA was then converted to cDNA with Superscript III Reverse Transcriptase (Invitrogen, 18080093) and oligo dT18 (NEB, S1316S) in the presence of RNase inhibitors. Diluted cDNA was assessed by qPCR using the Power SYBR Green PCR Master Mix (Thermo Fisher Scientific, 43-676-59) reagents, using technical duplicates. A standard curve generated from samples within each experiment was used to quantify relative amounts of transcript. Ifnb1 transcript levels (F, GTCCTCAACTGCTCTCCACT; R, CCTGCAACCACCACTCATTC) were normalized to housekeeping genes including Rps17 (F, CGCCATTATCCCCAGCAAG; R, TGTCGGGATCCACCTCAATG), Oaz1 (F, GTGGTGGCCTCTACATCGAG; R, AGCAGATGAAAACGTGGTCAG) or Hprt1 (F, GTTGGATACAGGCCAGACTTTGTTG; R, GAGGGTAGGCTGGCCTATAGGCT) as indicated in the figures. Human IFNB1 transcript quantities (F, CAGCATCTGCTGGTTGAAGA; R, CATTACCTGAAGGCCAAGGA) were normalized to HPRT1 (F, ATCAGACTGAAGAGCTATTGTAATGA; R, TGGCTTATATCCAACACTTCGTG). DNase-treated RNA that was not treated with reverse transcriptase was also included in qPCR assays as a control to ensure complete digestion of genomic DNA. RT–qPCR reaction wells with a poor ROX reference were excluded from analysis, as were samples with unusually low housekeeping gene amounts indicative of RNA degradation. Replicates in figures indicate biological replicates (separate wells of cells).
Roadblock RT–qPCR
BMMs were either pretreated with freshly prepared 400 μM 4SU (Cayman Chemicals, 16373) 1–2 h before 100 μg ml−1 DMXAA stimulation or treated with 4SU 2 h after DMXAA stimulation, as previously described27,62. In brief, RNA was collected as described above and DNase treated. Equal amounts of RNA (~200 ng or more) were treated with 48 μM N-ethyl maleimide (Sigma-Aldrich, 04259-5G) and quenched with 20 mM DTT. RNA was then purified with RNAClean beads (Beckman Coulter, A63987) and converted to cDNA with ProtoScript II Reverse Transcriptase according to the manufacturer’s instructions in the presence of RNase inhibitors. qPCR was then performed as described above with the primers for Ifnb1 (F, TGGATGGCAAAGGCAGTGTAA; R, CACCTACAGGGCGGACTTC) and Rps17 (see above). In some experiments, housekeeping-normalized Ifnb1 is displayed in the figures as the percentage of average Ifnb1/Rps17 quantity present at 2 h of DMXAA stimulation for each condition. To validate the Roadblock RT–qPCR protocol, in every Roadblock RT–qPCR experiment, BMM cells were also pretreated in parallel with 4SU before DMXAA treatment, and RNA was isolated at T = 2 and other timepoints indicated in the figures and converted to cDNA. NEM treatment was confirmed to reduce the detection of Ifnb1 transcripts by qPCR by around tenfold with 4SU pretreatment before DMXAA for every Roadblock RT–qPCR experiment.
ELISA
BMMs were seeded at 85,000 cells per well in non-tissue-culture-treated 96-well plates in 200 μl of medium and rested overnight. Cells were stimulated with stimuli indicated in the figure legends for 24 h. The plates were spun at 600g for 5 min, and the supernatants were removed and stored at −80 °C. Culture supernatants from DMXAA-treated cells were diluted 1:50–1:100 before evaluation using the Lumikine Xpress mIFN-β 2.0 kit according to the manufacturer’s instructions.
RNA-seq sample generation and analysis
Day 6 BMMs derived from B6 and Sp140−/−, or Ifnar−/− and Sp140−/−Ifnar−/− mice were seeded at 1 × 106 cells per well in six-well non-tissue-culture-treated plates and rested overnight. BMMs were then either left unstimulated or stimulated with DMXAA (100 μg ml−1 for B6 and Sp140−/− BMMs, and 10 μg ml−1 DMXAA for Ifnar−/− and Sp140−/−Ifnar−/− BMMs) for either 8 h for B6 and Sp140−/− BMMs or for 4 h for Ifnar−/− and Sp140−/−Ifnar−/− BMMs. RNA was isolated using the Omega Biotek Total RNA II kit, and DNase-treated with TURBO DNase (Thermo Fisher Scientific, AM2238) according to the manufacturer’s instructions. For RNA from B6 and Sp140−/− BMMs, single-index libraries were generated by Azenta, using rRNA depletion (Qiagen, QIAseq FastSelect–HMR rRNA Removal kit) and the NEBNext RNA Ultra kit (NEB). Libraries were evenly split across two lanes of an Illumina HiSeq flow cell and sequenced (150 bp paired-end reads, depth of 25–30 million reads per sample). For Ifnar−/− and Sp140−/−Ifnar−/− BMM samples, libraries were prepared for Illumina sequencing with dual indices using poly(A) selection and KAPA HyperPrep reagents (Roche) after heat fragmentation through the UC Berkeley QB3 Vincent J. Coates Genomics Sequencing Laboratory. Libraries were subsequently size-selected between 450 and 500 bp, and then sequenced on the Illumina NovaSeq flow cell for a depth of over 20 million mapped reads (150 bp, paired-end). B6 and Sp140−/− samples were collected separately from Ifnar−/− and Sp140−/−Ifnar−/− samples. Three biological replicates for each genotype and condition were collected across three independent experiments from three separate mice for each genotype, which were age and sex-matched. Adapters and low-quality reads were trimmed using BBDuk v.38.05 with arguments ‘ktrim=r k=23 mink=11 hdist=1 mapq=10 qtrim=r trimq=10 tpe tbo’. For UCSC genome browser visualization, reads were mapped to the mm10 mouse reference genome (https://genome.ucsc.edu/cgi-bin/hgGateway?db=mm10) using hisat2 v.2.1.0 with the options ‘–no-softclip -k 100 | samtools view -q 10 -Sb – | samtools sort’. CPM normalized bigwigs were made using deepTools bamCoverage v.3.0.1. For transcript quantification, reads were mapped to mm10 (gencode.vM18.annotation.gtf) using Salmon v.0.13.1 with the options ‘–libType A –validateMappings –rangeFactorizationBins 4 –gcBias’. DEGs were called using DESeq2 v.1.38.3 with design ‘~batch + genotype + treatment + genotype:treatment’. For differential expression analysis of Sp140−/− versus B6 BMMs and Sp140−/−Ifnar−/− versus Ifnar−/− BMMs, normalized count data were derived from the following DESeq2 comparisons: (1) SP140-deficient BMMs treated with DMXAA (three replicates) versus SP140-WT BMMs treated with DMXAA (3 replicates); and (2) untreated SP140-deficient BMMs (3 replicates) versus untreated SP140-WT BMMs (3 replicates). Genes with zero counts across all samples were removed. Volcano plots were generated with ggplot2 v.3.5.0 R package.
ATAC–seq sample generation and analysis
B6 and Sp140−/− samples BMMs were derived and stimulated as described above with 100 μg ml−1 DMXAA for 8 h in three separate experiments, in parallel to samples generated for RNA-seq. BMMs were collected in PBS and counted, and ATAC–seq samples were generated from 100,000 input cells essentially as described previously63, except that isolated nuclei were centrifuged at 1,000g. Illumina-compatible libraries were prepared as described previously63, with additional Ampure XP bead (Beckman Coulter) purification to remove contaminating adaptor dimers. The samples were sequenced with Azenta on the Illumina HiSeq flow cell (over 50 million paired end reads per sample, 150 bp reads). Adapters and low-quality reads were trimmed using BBDuk v.38.05 with the arguments ‘ktrim=r k=23 mink=11 hdist=1 maq=10 qtrim=r trimq=10 tpe tbo’ and mapped to mm10 using BWA-MEM v.0.7.15, and only uniquely mapping reads with a minimum MAPQ of 10 were retained. Fragments aligning to the mitochondrial genome were removed. Peak calling was performed using complete and size-subsetted alignment files with MACS2 v.2.1.1 with paired-end options ‘–format BAMPE –SPMR -B –broad’. For visualization, counts-per-million-normalized bigwig files were made using deepTools bamCoverage v.3.0.1. For differential expression analysis of ATAC–seq peak data of Sp140−/− BMMs treated with DMXAA (3 replicates) versus B6 BMMs treated with DMXAA (3 replicates), normalized count data were derived from DESeq2. The nearest gene was found to the resultant peaks in data generated from Sp140−/− BMMs using closestBed (v.2.28.0). This gene list was then overlapped with the closestBed gene list of HA–SP140 CUT&RUN peak data. Volcano plots were generated with ggplot2 v.3.5.0 R package.
BMM transduction
Low-passage HEK293T cells or GP2 packaging cells were seeded in a six-well format on tissue-culture-treated plates and rested at least overnight. To generate lentivirus, HEK293T cells at >70% confluency in a six-well tissue-culture-treated plate were transfected with 0.468 μg VSV-G (pMD2.G, Addgene, 12259), 1.17 μg D8.9 packaging vector and 1.56 μg of doxycycline-inducible, puromycin-selectable lentiviral vector per well using Lipofectamine 2000 according to the manufacturer’s instructions. The lentiviral backbone used in this study (pLIP) was adapted from pLIX (Addgene, 41394) by removal of ccdB29. Gene blocks encoding codon-optimized mouse/human RESIST, or mCherry were cloned into pLIP, digested with NheI and BamHI, after the dox-inducible promoter, using Infusion reagents (Takara), essentially as described previously29. RESIST(∆C) constructs were also generated by PCR to remove residues after Asp161 followed by Infusion cloning into the pLIP backbone. All of the constructs were validated by Sanger sequencing, the results of which were evaluated in Snapgene v.7.0.1. To generate retrovirus, GP2s at higher than 70% confluency in a six-well plate were transfected with 0.5 μg of VSV-G and 3.5 μg of retroviral vector per well with Lipofectamine 2000. Retroviral vectors in this study (SINV HA-SP140, SINV-SP140) were derived from the self-inactivating retrovirus pTGMP (Addgene, 32716; from the laboratory of S. Lowe). Mouse Sp140 or HA-Sp140 codon-optimized cDNA was cloned into pTGMP with Infusion (Takara), modified to include a minimal CMV promoter driving SP140 constructs, followed by a PGK promoter driving a puromycin-resistance cassette. Then, 18–20 h after transfection, medium on transfected cells was changed to 1 ml BMM medium. Bone marrow was collected as described above, and plated in BMM medium without dilution. Retronectin-treated (Takara) six-well plates were generated according to the manufacturer’s instructions. The next day (around 30 h after changing medium on transfected cells), virus was collected from transfected cells by filtration of supernatant through a 0.45 μm filter and added to 1 × 106 cells per well of bone marrow in 4 ml total of BMM medium. Plates were spun at 650g for 1.5–2 h at 37 °C. Then, 3 days after bone marrow collection, BMMs were fed with 1.33 ml BMM medium. BMMs were puromycin selected on day 4 after BMM collection with 2.75–5 μg ml−1 puromycin. Puromycin kill curves were determined for every stock to identify the lowest concentration needed for BMM selection, and a non-transduced well or BMMs transduced with a retroviral vector lacking a puromycin resistance cassette were used to verify complete killing of non-transduced cells by puromycin. After puromycin selection (2 days), the medium was exchanged and BMMs were allowed to recover for 2–6 days before seeding. BMMs transduced with lentiviral pLIP constructs were pretreated overnight for 24 h with 2.5 μg ml−1 doxycycline, then restimulated with 100 μg ml−1 DMXAA and fresh doxycycline. Cells were collected at the timepoints indicated in the figure legends for either RNA isolation or co-IP.
HA–SP140 CUT&RUN and analysis
Sp140−/− BMMs were transduced with retrovirus encoding HA–SP140 or SP140 as described above, and stimulated for 8 h with 100 μg ml−1 DMXAA. Half a million cells were input into CUT&RUN64 in biological triplicates using the Epicypher CUTANA ChIC/CUT&RUN kit (Epicypher, 14-1048, v3), using 0.5 μg of rabbit anti-HA monoclonal antibody (Cell Signaling Technologies, C29F4) with Escherichia coli genomic DNA spike-in. Non-transduced B6 BMMs (0.5 × 106) were also processed for CUT&RUN with 0.5 μg of rabbit isotype control IgG (Epicypher, 13-0042), with a single biological replicate. CUT&RUN was carried out on isolated nuclei according to the kit instructions. Libraries were prepared using the Epicypher CUTANA CUT&RUN library prep kit (Epicypher, 14-1001) according to the kit instructions, then sequenced on the Illumina NovaSeq flow cell with 250 bp paired-end reads for around 6 million reads per sample. Adapters and low-quality reads were trimmed using BBDuk v.38.05 using the options ‘ktrim=r k=23 mink=11 hdist=1 maq=10 tpe tbo qtrim=r trimq=10’. Trimmed reads were aligned to the mm10 assembly using BWA-MEM v.0.7.15, and only uniquely mapping reads with a minimum MAPQ of 10 were retained. Fragments aligning to the mitochondrial genome were removed. Peak calling was performed using complete and size subsetted alignment files with MACS2 v.2.1.1 with the paired-end options ‘–format BAMPE –pvalue 0.01 –SPMR -B –call-summits’. Bigwig files were prepared from the MACS2-normalized bedgraph files using bedGraphToBigWig v.4. MACS2 peak scores, the normalized number of sequence reads that originate from a bound genomic location, were output for HA–-SP140 peaks.
Cistrome and GREAT analysis
Replicate MACS2 CUT&RUN peak files were merged, then controls (IgG and SP140) were subtracted from the HA–SP140 peak file using bedtools intersect (v.2.28.0)65 to output a file of SP140 peaks. This SP140 peak file was used as input in the cistrome toolkit data browser (http://dbtoolkit.cistrome.org/)66 looking for significant binding overlap of histone marks and variants in mm10. The SP140 peak file was also used as input for the Genomic Regions Enrichment of Annotations Tool (GREAT, v.4.0.4)67.
Recombinant protein expression and purification
ANXA2-S100A was produced and purified in E. coli BL21 (DE3) Star cells (Thermo Fisher Scientific) in LB medium at 20 °C as a fusion protein carrying an N-terminal His6–SUMO tag. Cells were resuspended in lysis buffer (50 mM HEPES, 500 mM NaCl, 25 mM imidazole, pH 7.5) and lysed using the Branson Ultrasonics Sonifier SFX550. The lysate was cleared by centrifugation at 40,000g for 1 h at 4 °C. The cleared lysates were loaded onto the 5 ml HisTrap column (Cytiva). The bound protein was eluted over a linear gradient with elution buffer (50 mM HEPES, 200 mM NaCl, 500 mM imidazole, pH 7.5). For the final step, size-exclusion chromatography was performed on the Superdex 200 26/600 column in a buffer containing 10 mM HEPES, 200 mM NaCl, 2 mM DTT, pH 7.5.
For expression of full-length RESIST, full-length human RESIST (UniProt: Q3ZCQ2) was inserted between the BamHI and XbaI restriction sites of the pLIB plasmid68 with a TEV (tobacco etch virus) protease-cleavable, N-terminal His6–MBP (maltose-binding protein) tag and a C-terminal StrepII tag. The DNA sequence encoding the RNA-binding zinc fingers of human TTP (TZF; UniProt: P26651, residues Ser102 to Ser169) was inserted between the NdeI and XhoI restriction sites of the pnYC plasmid69 with a TEV-cleavable N-terminal MBP tag and a C-terminal StrepII tag. DNA constructs for the expression of the NOT9 module were previously described70. Subsequently, full-length human RESIST with an N-terminal TEV-cleavable His6-MBP tag and a C-terminal StrepII tag was expressed in Sf21 insect cells using the MultiBac baculovirus expression system71,72 as previously described73. In brief, Sf21 cells were grown to a density of 2 × 106 cells per ml at 27 °C in Sf900II medium (Thermo Fisher Scientific), infected with the V1 generation His6–MBP–RESIST–StrepII baculovirus, and collected 48 h after they stopped dividing. The collected cells were resuspended in ice-cold protein buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 5% (v/v) glycerol, 20 mM CHAPS, 25 mM imidazole) and lysed by sonication. The lysate was clarified by centrifugation at 40,000g for 40 min at 4 °C, filtered through a 0.45 μm nylon filter and loaded onto a 5 ml nickel-charged HisTrap column (Cytiva). Contaminants were removed by washing with lysis buffer supplemented with 40 mM imidazole, and His6–MBP–RESIST–StrepII was eluted in lysis buffer supplemented with 250 mM imidazole. The eluted protein was further purified by size-exclusion chromatography on the HiLoad Superdex 200 16/600 column (Cytiva) in buffer containing 50 mM HEPES pH 7.5, 150 mM NaCl, 5% (v/v) glycerol, 20 mM CHAPS. The peak fractions were pooled, concentrated with a centrifugal filter, flash-frozen in liquid nitrogen and stored at −80 °C. The RNA-binding zinc fingers of TTP (TZF) were expressed in E. coli BL21(DE3) Star cells (Thermo Fisher Scientific) in autoinduction medium74 at 20 °C overnight as a fusion protein carrying an N-terminal, TEV-cleavable MBP tag and a C-terminal StrepII tag. Collected cells were resuspended in protein buffer (50 mM HEPES pH 7.5, 300 mM NaCl, 10% (w/v) sucrose) and lysed by sonication. The lysates were clarified by centrifugation at 40,000g for 40 min and loaded onto the 1 ml StrepTrap XT column (Cytiva). Contaminants were removed by washing with high-salt buffer (50 mM HEPES pH 7.5, 1 M NaCl, 10% (w/v) sucrose) before elution with lysis buffer supplemented with 50 mM biotin. Eluted protein was further purified by size-exclusion chromatography on the Superdex 200 26/600 column (Cytiva) in protein buffer supplemented with 2 mM DTT. The peak fractions were then pooled, concentrated with a centrifugal filter, flash-frozen in liquid nitrogen and stored at −80 °C. Finally, The NOT9 module was prepared as previously described48.
StrepTactin pull-down assay
StrepII-tagged MBP, as well as StrepII-tagged and SUMO-tagged SMARCA3 (residues 26–39) were produced in E. coli BL21 (DE3) Star cells (Thermo Fisher Scientific) grown in autoinduction medium overnight at 37 °C. Cells were resuspended in lysis buffer (50 mM HEPES, 500 mM NaCl, pH 7.5) and lysed using the Branson Ultrasonics Sonifier SFX550, the lysate was then cleared by centrifugation at 40,000g for 1 h at 4 °C. StrepII-tagged proteins (ANXA2R, MBP or SMARCA3) were incubated with StrepTactin Sepharose resin (Cytiva, 28935599). After incubation for 1 h, the beads were washed twice with 50 mM HEPES, 500 mM NaCl, pH 7.5, 0.03% Tween-20, once with 50 mM HEPES, 500 mM NaCl, pH 7.5 and once with binding buffer (50 mM HEPES, 200 mM NaCl, pH 7.5). Purified ANXA2(S100A) was added to the bead-bound proteins. After incubation for 1 h, the beads were washed four times with binding buffer and proteins were eluted with 50 mM biotin in binding buffer. The eluted proteins were analysed by SDS–PAGE followed by Coomassie blue staining.
For pull-downs of full-length human RESIST with CCR4–NOT subunits, purified His6–MBP–RESIST–StrepII or His6–MBP–StrepII were immobilized as bait through the C-terminal StrepII tag on streptavidin agarose resin prepared in-house. Then, 250 pmol of bait protein was incubated for 1 h in pull-down buffer (50 mM HEPES pH 7.5, 200 mM NaCl, 0.03% (v/v) Tween-20) at 6 °C under constant agitation. Unbound protein was removed after two washes with pull-down buffer, and 500 pmol of NOT9 module was incubated for 1 h with the bead-bound protein. Finally, the beads were washed three times with a pull-down buffer, and the bound proteins were eluted using a pull-down buffer supplemented with 50 mM biotin. The eluted proteins were analysed using SDS–PAGE followed by Coomassie blue staining.
BlaER1 transduction and stimulation
Lentivirus was generated from HEK293T cells transfected with pLIP constructs (mCherry or human RESIST, either untagged or with N/C-terminal HA tags) as described for BMM transduction above. Virus was overlaid onto BlaER1 cells, which were subsequently puromycin selected and differentiated as described previously29. After 5 days of differentiation, BlaER1 cells were stimulated with 2.5 μg ml−1 doxycycline and fresh cytokines overnight. Medium with new cytokines, fresh doxycycline and ADU-S100 at a final concentration of 5 μg ml−1 (Aduro) was added the next day. At the indicated timepoints, adherent and non-adherent cells were collected and lysed in TRK lysis buffer (Omega Biotek Total RNA kit) for RNA isolation. RNA was isolated as quickly as possible from lysates as described above, and converted to cDNA with Superscript reagents and RNase inhibitors as described above.
Co-IP analysis
For IP analysis of RESIST from BMMs, BMMs were transduced with RESIST constructs or mCherry and treated with doxycycline, followed by DMXAA treatment, as described above. Cells (0.6 × 106–2.4 × 106) were collected at the timepoints indicated in figure legends, and lysed on ice in 300–600 μl lysis buffer (50 mM Tris-HCl pH 7.5, 0.2% NP-40, 5% glycerol, 100 mM NaCl, 1.5 mM MgCl2, 1× protease inhibitor cocktail) for 30 min. The lysates were clarified by centrifugation at 18,213g for 30 min at 4 °C and quantified using the bicinchoninic acid assay to ensure approximately equal amounts of input protein. One tenth of clarified lysate was diluted in Laemmli buffer for input sample. Supernatant was incubated with anti-HA magnetic beads (Thermo Fisher Scientific, 88836) with rotation for 3 h at 4 °C. Beads with lysates were then washed three times with lysis buffer (1 ml per wash), then immunoprecipitated proteins were eluted in 30–50 ml Laemmli buffer by boiling for 5 min. The samples were then analysed by immunoblotting.
For IP analysis of Flag–TTP from BMMs75, N-terminally Flag-tagged mouse TTP constructs were cloned into pLIP as described above. Mouse Flag-TTP was transfected into semi-confluent 10 cm plates of HEK293T cells with N-terminally HA-tagged mouse RESIST or an equivalent amount of mCherry using Lipofectamine 2000 according to the manufacturer’s instructions. 600 ng of TTP construct was co-transfected with 6 μg mCherry, and 660 ng of TTP construct was co-transfected with 6 μg RESIST. After transfection, cells were treated with doxycycline to induce expression for 24 h, then collected in PBS. Cells were lysed in hypotonic lysis buffer consisting of 10 mM Tris-HCl pH 7.5, 10 mM NaCl, 2 mM EDTA, 0.5% Triton X-100, and protease inhibitors for 10 min on ice with vortexing. NaCl was then adjusted to 150 mM and the samples were treated with 30 μg of RNase A on ice for 20 min with vortexing. The samples were clarified by centrifugation at 18,000g for 30 min at 4 °C, and 10% of the sample was taken as the input sample and diluted in Laemmli buffer. The sample was incubated with 50 μl of Sigma anti-flag M2 agarose beads (Sigma Aldrich, M8823) equilibrated in lysis buffer with adjusted NaCl and resuspended in a total volume of 200 μl lysis buffer with adjusted NaCl per reaction. The samples were incubated with beads for 2 h with rotation at 4 °C, then washed five times with 1 ml wash buffer (50 mM Tris HCl pH 7.5, 300 mM NaCl, 0.05% Triton X-100, protease inhibitors). The remaining liquid was completely aspirated from the beads with a small-bore needle, and the beads were resuspended in 2× Laemmli buffer diluted in wash buffer before analysis by immunoblot.
Immunoblot
Samples diluted in Laemmli buffer were run on 4–12% Bis-Tris protein gels (Invitrogen) and then transferred to PVDF membranes at 35 V for 90 min. Membranes were blocked in either Odyssey Licor PBS blocking buffer or in 2–5% non-fat dry milk diluted in TBST. The membranes were then probed with antibodies at 4 °C overnight diluted in 5% BSA TBST. The antibodies used in this study were as follows: rat anti-HA (Roche, 3F10, 1186742300, 1:1,000), mouse anti-actin (Santa Cruz Biotechnology, sc-47778, 1:1,000), rabbit anti-CNOT1 (Cell Signaling Technologies, 44613S, 1:1,000), rabbit anti-CNOT9 (Proteintech, 22503-1-AP, 1:500), rabbit anti-TTP (Millipore Sigma, ABE285, 1:1,000), rabbit anti-CNOT11 (Sigma-Aldrich, HPA069823, 0.4 μg ml−1), rabbit anti-ZFP36L1 (Cell Signaling Technologies, 30894S, 1:1,000), rabbit anti-ZFP36L2 (Abcam, ab70775, 1:1,000), and rabbit anti-SP140 (Covance, as previously described4; 1:1,000) and rabbit anti-Flag for Flag–TTP IPs (Thermo Fisher Scientific, PA1-984B, 1:1,000).
In vivo L. pneumophila infections
L. pneumophila infections were performed as described previously4. In brief, JR32ΔflaA L. pneumophila (from the laboratory of D. Zamboni) was streaked from a frozen glycerol stock onto BCYE plates. A single colony was used to streak an approximately 4 cm2 patch that was subsequently grown for 2–3 days. Bacteria were diluted in water and the optical density was measured at 600 nm to determine the bacterial concentration. Bacteria were then diluted to a final concentration of 2.5 × 106 bacteria per ml in sterile PBS. Mice were anaesthetized with a mixture of xylazine and ketamine through intraperitoneal injection, then 40 μl of diluted bacteria was administered intranasally (final infectious dose of 105 bacteria per mouse). At 96 h after infection, mice were euthanized and the lungs were homogenized in 5 ml of autoclaved MilliQ water. Lung homogenate was diluted and plated on BCYE plates, and colony-forming units were enumerated after 4 days of growth. One infection of Sp140−/−Resist1/2+/+ and Sp140−/−Resist1/2−/− littermates was performed, which reproduced the results obtained with non-littermate infections.
AlphaFold structure predictions
AlphaFold-Multimer (v.2.3.2)76 was run on equipment hosted by the Cal Cryo EM facility comprising an Nvidia GPU and >72 TB of storage space. Mouse RESIST and CCR4–NOT amino acid sequences were from NCBI (RESIST, XP_006517870.1; CNOT1 M-HEAT, XP_036009857.1, with a start codon followed by residues 815–1007; CNOT11, NP_082319.1; CNOT1 N-MIF4G, XP_036009857.1, residues 1–695; CNOT10, NP_705813.2; CNOT9, NP_067358.1). In brief, AlphaFold was run in multimer mode on RESIST with CNOT1 M-HEAT, CNOT9 or CNOT1 N-MIF4G/CNOT10/CNOT11 with the default settings, the max template date specified as 2023-01-01 and –db_preset=full_dbs. Output models were visualized in ChimeraX (v.1.6.1), and aligned using the matchmaker command. PAE plots and structures coloured by pLDDT were visualized with PAEViewer (v.1.0)77.
Gene disruption in BMMs with Cas9–gRNA electroporation
BMMs were collected in PBS and electroporated with Cas9 2 NLS nuclease (Synthego) complexed with gRNAs (Synthego, sgRNA EZ kits) and Alt-R Cas9 Electroporation Enhancer (IDT, 1075916) in Lonza P3 buffer (Lonza, V4XP-3032) with buffer supplement as described previously78. Electroporation was performed using the Lonza 4D-Nucleofector Core Unit (AAF-1002B) using the program CM-137. Electroporated BMMs were immediately plated in BMM medium. A half-medium exchange was performed every 2 days until day 10 after bone marrow collection, when BMMs were seeded for downstream assays. The knockout efficiency was evaluated by immunoblotting or PCR analysis of genomic DNA for targeted regions followed by ICE analysis (Synthego) as indicated in figure legends. Gm21188 (Resist1) was genotyped with Primestar PCR reagents and the primers F1 (ATTGAGAAATCCGTTTGTAATGGG) and R1 (TAGGCGAATTTCGTGGCACA) according to the manufacturer’s instructions with an annealing temperature of 55 °C, or using Q5 PCR reagents and primers F2 (TTGAGAAATCCGTTTGTAATGGG) and R2 (GCCTTTCTCCGGATTCACGA) with the cycling conditions: 98 °C for 3 min; 35 cycles of 98 °C for 10 s, 63.5 °C for 20 s and 72 °C for 1 min 5 s. Gm36079 (Resist2) was genotyped with F (TGGTATTCTCTAGAGATAACATCACAGCACCTACTTACTCC) and R (CCTCCCCTCGCCATCACTGCCTG) using the Phusion GC PCR reagents and cycling conditions of: 98 °C for 30 s, then 30–35 cycles of 98 °C for 10 s, 72 °C for 15 s, 72 °C for 1 min. Disruption of Rc3h1 was determined by PCR (F, CACACTATGTGCTGACTGTATCTACAGAAG; R, TCCCCTCAGGTAAAACAGTGC; cycling: 98 °C for 30 s, then 30 cycles of 98 °C for 10 s, 60 °C for 5 s and 72 °C for 1 min) with Phusion GC PCR reagents. Disruption of Rc3h2 was determined by PCR (Q5 PCR reagents with F2, AGGGCATAAGATGTTGCACAGA; R2, ACTGCTAACCCGAGCATCAG; and cycling of 98 °C for 3 min, then 35 cycles of 98 °C for 10 s, 60 °C for 20 s and 72 °C for 40 s). PCRs were cleaned by gel extraction, Ampure XP beads (Beckman Coulter) or treatment with FAST-AP and ExoI before submission for Sanger sequencing. ICE analysis was performed with the Synthego ICE online tool (https://ice.editco.bio/#/). The gRNA sequences used in this study were as follows: Gm21188/Gm36079 gRNA 1, GCUGGGCCUCUUGCACCAGA; Gm21188/Gm36079 gRNA 2, CGACGAUGGCGGUGACUACC; Cnot1 gRNA 1, UGUGAAUCGGCACGGUCCUG; Cnot1 gRNA 2, ACUCAUUCAGGAUUAACAGA; Cnot11 gRNA 1, UCCAUCAAGGCAAUCUGGCG; Cnot11 gRNA 2, GCUGAGCAUCAUCUCGGAGG; Cnot9 gRNA 1, CAUUGCAAACUCUGUUAGAC; Cnot9 gRNA 2, GCCUACUGCACUAGCCCAAG; Zfp36 gRNA 1, CAUGACCUGUCAUCCGACCA; Zfp36 gRNA 2, CUUCAUCCACAACCCCACCG; Zfp36l1 gRNA 1, AAAAAUGGUGGCGGACACGA; Zfp36l1 gRNA 2, ACGGGCAAAAGCCGAUGGTG; Zfp36l2 gRNA 1, CAAGAAGUCGAUAUCGUAGA; Zfp36l2 gRNA 2, GAGAGCGGCACGUGCAAGUA; Rc3h1 gRNA, CAAAUGGGCAAGCCUUACGG; Rc3h2 gRNA, UCGGUGAAGUUUAUUCAAGC.
TTP EMSA
The substrate RNAs were generated by in vitro transcription (IVT). For the IFNB1 WT RNA substrate, the ARE in the 3′-UTR of the IFNB1 mRNA (GenBank: NM_002176.4; nucleotides 740–825) was synthesized as a gene fragment (Azenta) with an upstream T7 promoter and 17 random nucleotides downstream. All adenosine residues between nucleotides 758 and 825 were mutated to cytosine for the IFNB1-MUT RNA substrate. The gene fragments were amplified by PCR, and the purified PCR products were used as templates for IVT using the HiScribe T7 High Yield RNA Synthesis Kit (NEB). IVT products were separated by size-exclusion chromatography on the Superdex 200 increase 10/300 GL in buffer containing 10 mM HEPES pH 7.5, 200 mM NaCl. The fractions containing the intact RNA substrates were pooled, ethanol precipitated and resuspended in RNase-free water. Electrophoretic mobility shift assay (EMSA) binding reactions contained 50 nM substrate RNA and 50–800 nM TZF protein. The reactions were carried out for 15 min at 37 °C in a buffer containing 20 mM PIPES pH 6.8, 10 mM KCl, 40 mM NaCl, 2 mM Mg(OAc)2, 3% (v/v) Ficoll 400 and 0.05% (v/v) NP-40. The RNA–protein complexes were analysed by electrophoresis on a nondenaturing polyacrylamide gel in 0.5× TBE buffer, pH 8.3, at 10 V cm−1. Gels were stained in 0.5× TBE pH 7.5 with 1× SYBR Gold (Thermo Fisher Scientific) for 5 min before analysis. Images were quantified using FiJi79.
Viral infections of BMMs
Day 7 BMMs were generated as described above, and 250,000 cells were infected with viruses at the indicated multiplicities of infection in a non-tissue-culture-treated 12-well plate. For MCMV-GFP and MHV68-GFP, cells were infected for 3–4 h in serum-free RPMI supplemented with penicillin–streptomycin and glutamine in a low volume of inoculum. MCMV-GFP and MHV68-GFP were a gift from L. Coscoy and B. A. Glaunsinger. MHV68-GFP and MCMV-GFP titre was estimated by infection of 3T3 cells, and calculated by the assumption that a viral dilution resulting in 100% of infected 3T3 cells corresponds to an approximate multiplicity of infection of 5. For Sendai-GFP infections (ViraTree), cells were infected for 1.5 h in serum-free RPMI supplemented with penicillin–streptomycin and glutamine in a low inoculum volume. After infection, medium was replaced and BMMs were cultured for an additional 20–24 h before cells were collected in PBS, stained with Ghost Dye Far Red 780, fixed with the BD Cytofix Cytoperm kit according to the kit instructions, then washed and analysed by flow cytometry. Data were analysed using FlowJo.
Immunofluorescence microscopy
BMMs were seeded on glass coverslips between 0.5 × 106 and 1 × 106 per slip in BMM medium lacking antibiotics and rested overnight. BMMs were stimulated with 100 μg ml−1 DMXAA for 8 h, fixed in 4% freshly prepared paraformaldehyde (Electron Microscopy Sciences) for 10 min at room temperature, then permeabilized in freshly made 0.2% Triton X-100 and 0.2% BSA in PBS on ice for 10 min. The coverslips were washed three times with PBS and then blocked in goat serum and FC-block (TruStain FcX PLUS, anti-mouse CD16/32) for 1 h, then incubated overnight at 4 °C in primary antibody diluted in PBS with 1% Tween-20 and 1% BSA. Primary antibodies and dilutions were mouse anti-PML (Millipore Sigma, 05-718, 1:100), rat anti-HA (Roche, 11867423001, 1:200), rabbit anti-fibrillarin (Abcam, ab166630, 1:100). Coverslips were washed three times in PBS, then incubated in secondary antibody diluted 1:1,000 in PBS with 1% Tween-20 and 1% BSA for 2–3 h at room temperature. Secondary antibodies used were donkey anti-rat Alexa Fluor 488 (Invitrogen, A21208), goat anti-mouse Alexa Fluor 647 (Invitrogen, A21236), goat anti rabbit 647 (Life technologies, A21244). After three PBS washes of coverslips, nuclei were stained with DAPI at 1 μg ml−1 in PBS for 10 min at room temperature, followed by three PBS washes. The coverslips were mounted in Vectashield mounting medium (Vector Laboratories, H-1000-10). Coverslip edges were then sealed with clear nail polish before imaging on a Zeiss LSM 880 NLO AxioExaminer at ×63 magnification.
IF image processing and quantification
For independent experiments as indicated in figure legends, z stacks were processed by Imaris File Converter v.10.0.1 followed by Imaris Stitcher v.9.9.1. Images were Gaussian filtered (0.132 μm) and screenshots were generated for figures. Surfaces for DAPI, HA–SP140, PML and fibrillarin were generated in Imaris using split-touching of 1 μm for HA–SP140, PML and fibrillarin surfaces and 5 μm for DAPI surfaces. Surface statistics were then exported. Surfaces were first filtered as DAPI+ (within 2 s.d. below average fluorescence intensity of DAPI surfaces) and then filtered by size (over 0.2 μm3). HA–SP140 NBs were considered to overlap fibrillarin if the fibrillarin intensity mean for an HA–SP140 surface was within 2s.d. of the average fluorescence intensity for fibrillarin surfaces, and vice versa for fibrillarin surfaces overlapping with HA–SP140 surfaces. The same criteria for overlap were applied to HA–SP140 and PML surfaces. The mean fluorescence intensities of surfaces were calculated for over 100 nuclei for each independent experiment.
Statistics and reproducibility
All results except for HA–SP140 CUT&RUN and AlphaFold predictions were repeated at least twice in independent experiments. Statistics and graphs for all experiments except for RNA-seq, ATAC–seq and CUT&RUN experiments were generated using GraphPad Prism (v.10.0.2). For data with two groups of comparison, P values were calculated with two-tailed t-tests using Welch’s correction or two-way ANOVA, as described in the figure legends. For data with more than two comparison groups, ANOVA was used. We found that the residuals for our RT–qPCR data were not normally distributed and for these data we therefore performed ANOVA on log10-transformed data, which generated more normally distributed residuals based on Q–Q plots, therefore more appropriate for ANOVA tests. We used one-way Welch’s and Brown–Forsythe ANOVA without assuming data sphericity or equal variance, for data with multiple genotypes and one treatment condition, with a more-conservative post hoc Dunnett’s T3 multiple-comparison correction for log10-transformed data and less-conservative post hoc FDR correction for multiple comparisons (Q = 0.001, two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli) for all other data. Two-way ANOVA was performed for data with more than two comparison groups and/or multiple timepoints of measurement, with a full model including an interaction term, as we found that the effect of genotype varied across time. For two-way ANOVA, we did not assume sphericity, and used post hoc Tukey’s multiple-comparison test or Šidák’s multiple-comparison correction as described in the figure legends. For non-normally distributed data (L. pneumophila in vivo infections), Kruskal–Wallis one-way ANOVA with Dunn’s multiple-comparison test was used. The mean for all data was graphed, and replicates are individually represented by dots. Error bars indicate the s.e.m. Data shown in each figure represent the provided source raw data; statistical test details are also provided in the Source data. Replicates in RT–qPCR, ELISA or viral infections represent separate wells within an experiment, while replicates in L. pneumophila infections represent individual mice from three combined experiments. Replicate numbers (n) are represented in the figures, legends and Source data. For all experiments, samples were grouped based on genotype and treatment group and were not further randomized. Investigators were not blinded to experimental groups, and statistical methods were not used to predetermine sample size.
Schematics
All schematics were generated in BioRender by R.E.V. Schematics are available at https://BioRender.com/owrrzg2 for Fig. 4h, https://BioRender.com/0kcpnq3 and https://BioRender.com/rig1le4 for Fig. 5h, and https://BioRender.com/22n4jit for Extended Data Fig. 4a.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.