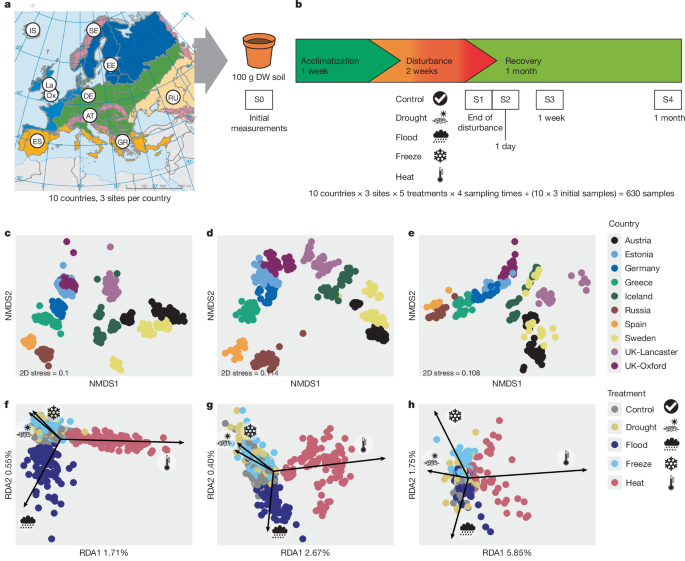
Study sites and soil sampling
We limited our study to natural or extensively managed grasslands across Europe, aiming to cover the widest range possible of climates and soil types. Our grassland network consisted of ten locations (referred to as countries) covering most biogeographical regions in Europe19 (Fig. 1a,b and Supplementary Table 1): alpine (AT, Austria), subarctic (SE, Sweden), Arctic (IS, Iceland), Atlantic (UK-Ox, Oxford and UK-La, Lancaster, both UK), boreal (EE, Estonia), continental (DE, Germany), Mediterranean (ES, Spain and GR, Greece) and Steppic (RU, Russia). Climatic data associated with the collection sites were obtained from the WorldClim database in April 2018 (ref. 46), based on observations recorded between 1960 and 1990 at 2.5âmin resolution, using the coordinates of the sites. We captured a wide range of MAP values, varying from 223âmm in Russia to 1,383âmm in Austria, whereas MAT varied between â2â°C in Sweden and 14.5â°C in Spain. Seasonal variability varied depending on the location, with Spain having the highest precipitation seasonality (38âmm) and UK-Oxford the lowest (14âmm). Russia varied the most in regard to temperature seasonality (38.5â°C) and Iceland the least (14â°C). We aimed to collect soil samples at peak microbial biomass, which is close to peak vegetation biomass, when the average temperature was the closest to 18â°Câthat is, in spring for southern locations and following snow melt, in summer, for northern locations. In May 2018 we collected soil from Russia, Greece, Spain and Estonia, followed by Germany and Oxford in June, Austria and Iceland in July and Lancaster and Sweden in August.
Three replicate sites per country were selected to encompass a wide range of soil types present in European grasslands, for a total of 30âreplicate sites, closely situated within countries (0.05â11.76âkm apart). According to the World Reference Base47 (https://soilgrids.org), our study included seven soil type categories, most of these belonging to the Haplic Cambisols (16), followed by Haplic Podzols (6), Haplic Kastanozems (3), Haplic Luvisols (2), Petric Calcisols (1), Rendzic Leptosols (1) and Haplic Gleysols (1). The most acidic soils were collected in Austria (pHâ4.73), with those from Spain having the highest pH (7.89). Within each replicate site, seven 1âÃâ1âm2 plots were arranged at least 5âm apart. We removed vegetation and stones and randomly sampled four soil cores from each plot with a 3-cm-diameter soil corer (0â15âcm). We carefully sterilized the collecting tools with ethanol 70% and distilled water and sampled the soil with gloves to avoid cross-contamination. Soil cores were pooled and sieved at 4âmm to form one homogenized composite soil sample per replicate site. Soil samples were transported in cold boxes to the laboratory and stored at 4â°C before analysis or to the establishment of microcosms. We measured WHC by immersing 100âg of fresh soil in water overnight and measuring the mass of water at saturation (100% WHC) following 4âh of drainage at 18â°C. We determined soil moisture content following drying at 105â°C for 48âh. This information was used to calculate the amount of fresh soil corresponding to 100âg of dry soil, and to maintain the moisture of the microcosms during the experiment.
Microcosm procedure and harvest
For each replicate site, fresh soil was thoroughly mixed and dispatched into 20âpots (240âml, 7.6âcm diameter across the top). We subsampled 20 and 5âg of soil for instant determination of initial soil moisture and nutrient concentrations, and stored 4âg of soil at â20â°C for DNA-based analyses. Considering the variability in soil density, WHC or initial moisture of soil derived from the different sites, the amount of soil per microcosm was standardized by dry weight (100âg dry weight per pot). Microcosms were placed for 7âdays in a Percival AR-66L2 growth chamber (CLF PlantClimatics), open to the air, at 18â°C to allow acclimatization of microbial communities and adjustment of soil moisture to 60% WHC with milliQ water. Following this period, microcosms were randomly divided into five treatments. Temperature and moisture parameters were manipulated as follows for 14âdays: control (18â°C, 60% WHC), drought (18â°C, 10% WHC), flood (18â°C, 100% WHC), freeze (â20â°C, 60% WHC) and heat (35â°C, 60% WHC). Application of these treatments for 2âweeks imposed extreme disturbance on soils from all locations usedâfor example, temperatures of â20â°C do not commonly occur even in Arctic soils48 whereas 35â°C is rarely exceeded in semiarid systems and during extreme heatwaves49,50. All treatments were placed in the same growth chamber and grouped by replicate site, where control exhibited the same environmental parameters as the acclimatization period: for drought we stopped watering the pots when moisture reached 10% WHC (Supplementary Fig. 4) and, for flood, microcosms were placed in plastic cups and submerged 1âcm above the soil surface. Freeze and heat samples were placed in a separate freezer and growth chamber, respectively, for the duration of the disturbance. At the end of the disturbance, one set of microcosms was harvested (see below) and the remainder returned to the main growth chamber, randomized within block (replicate site). Moisture content for drought was brought back to 60% WHC and, for flood, microcosms were drained until they reached the appropriate moisture content (Supplementary Fig. 4). To maintain moisture content at 60% WHC, the respective microcosms were watered by weight every 2âdays with milliQ water. We sacrificed one microcosm per treatment and per replicate site at four sampling times: end of disturbance (S1) and at 1 (S2), 8 (S3) and 26 (S4) days following the end of the disturbance (5âÃâ3â=â15 pots per harvest). This resulted in a total of 600âmicrocosms: 10âcountriesâÃâ3 replicate sitesâÃâ5 treatmentsâÃâ4 sampling times. Ten Russian samples at S2 were excluded from the study because of insufficient soil amount.
At each sampling time, we measured the emission of greenhouse gases in the dark by placing the microcosms into 500âml airtight Kilner jars covered in aluminium foil. The jars were sealed and 10âml gas samples taken sequentially in the headspace with a syringe at 0, 10, 20 and 30âmin. Gas samples were transferred to 20âml pre-evacuated glass vials, and CO2, N2O and CH4 concentrations analysed by gas chromatography (GC Agilentâ7890B). All gas concentrations were converted into fluxes based on soil dry weight (microgram of gas per gram of dry soil per hour). Fluxes were calculated considering the variation in both headspace volume (soils have different bulk densities depending on their origin, and headspace gas volume decreased by gas sampling) and temperature at each measurement. Microcosms were then thoroughly homogenized, with immediate subsampling of 20 and 5âg to determine soil moisture and nutrient concentrations. In addition, we stored 52âg (40â+â(6âÃâ2âg)) of soil at â20â°C for DNA-based analyses, MicroResp and enzymatic profiles. DNA was extracted within 3âmonths following freezing.
Soil analyses and microbial activity
Dissolved carbon and nitrogen in initial soil samples (nâ=â30) and all microcosms (nâ=â590, 600âââ10 Russian samples as described above) were extracted from 5âg of soil (fresh weight) in 35âml of milliQ water, shaken for 10âmin at 220ârpm and centrifuged for 15âmin at 4,000ârpm. Water extracts were filtered through cellulose papers (Whatman no.â1, 11âµm) and syringe filters (0.45âµm), and filtrates were stored at 4â°C for a maximum of 15âdays. Filtrates were analysed on a Seal AutoAnalyzer3 Segmented Flow Multi-chemistry analyser (Mequon) to measure dissolved nitrogen. Dissolved organic nitrogen was obtained by subtracting total dissolved inorganicâN (NH4+-N and NO3â-N) from total dissolved N. Dissolved organic carbon was obtained from the same filtrates analysed on a 5000A TOC analyser (Shimadzu). Soil pH was measured on initial samples (30âsamples) by the water method using a volume ratio of 2:5 fresh soil:milliQ water. Total soil C and N were determined by highâtemperature combustion (150âmg of ovenâdried, ground subsamples), followed by thermoconductometric detection on a Vario EL cube (Elementar Analysensysteme).
Substrate-induced respiration profiles were measured by the MicroResp method51 on soil samples collected at S1 and S4 (280âsamples). Soils were defrosted overnight at 4â°C. We used the microplate filler device (300âµl) to standardize the volume of soil added in each well of the deep-well microplate (1.2âml) and recorded the mass of soil added. We prepared detection microplates with a pH indicator gel to estimate the amount of CO2 produced by a colourimetric method (3% agar, 2.5âmM NaHCO3, 150âmM KCl and 12.5âµgâmlâ1 cresol red). We dispensed 25âµl of eight substrate solutions relevant to soil microbial activity separately to the wells: water, glucose, fructose, carboxymethyl cellulose, citric acid, malic acid, alanine or asparagine. Carbon substrates were prepared to a concentration of 12âmgâCâgâ1âsoil/water, corresponding on average to 30âmgâglucoseâgâ1âsoil/water solution. Initial absorbance of the detection plates was determined at 570ânm on a Clario Star microplate reader. We incubated the sealed system deep-well microplate/detection plate for 6âh at 25â°C in the dark before measurement of final absorbance. Respiration rates (microgram of CO2-C per gram of dry soil per hour) were calculated according to ref. 51.
Enzymatic activity assays were performed on soil samples at S1 and S4. Microbial potential enzymatic activities were assessed for acetylesterase, β-glucosidase, phosphatase and leucine-aminopeptidase, with methylumbelliferyl- and 7-amino-4-methylcoumarin-conjugated substrates (Sigma-Aldrich), using the protocol described in ref. 52. In brief, a total of 1.5âg of frozen soil was mixed with 20âml of milliQ water and shaken for 20âmin at 400ârpm. We added 30âµl of soil solution to a microplate containing 170âμl of substrate solution at 300âmM, and incubated it at 28â°C for 3âh.
DNA extraction and sequence processing
We extracted total DNA from 250âmg of frozen soil from the initial samples (nâ=â30) and microcosm samples (nâ=â600âââ10) using the DNeasy PowerSoil Pro Kit, following the manufacturerâs instructions (Qiagen). Tubes were vortexed for 10âmin at 1,200ârpm on a FastPrep-96 instrument (MP Biomedicals) at maximum speed, and DNA was eluted in 80âµl of the elution buffer. DNA integrity and quality were validated by running DNA samples on a 1.5% agarose gel at 75âV for 55âmin, and by checking 280:260 absorbance ratios with the Clario Star microplate reader (BMG LABTECH). We quantified DNA concentrations by fluorimetry using the Quant-iT dsDNA Broad-Range Assay Kit (Life technologies) with the Clario Star microplate reader. DNA solutions were split among three tubes and stored at â20â°C until use for whole-metagenomic sequencing and prokaryotic and fungal metabarcoding.
Prokaryotic and fungal community composition of the initial and microcosm samples at all sampling times (nâ=â620) was assessed by sequencing the V4â5 region of 16S rRNA genes using the primers 515âforward GTGYCAGCMGCCGCGGTAA and 806âreverse GGACTACNVGGGTWTCTAAT53, and established primers fITS7 (GTGARTCATCGAATCTTTG) and ITS4 (TCCTCCGCTTATTGATATGC) coding the ITS2 region54, respectively. We followed the PCR protocols of the Earth Microbiome Project55, and sequencing was performed using a two-step Nextera approach on the Illumina MiSeq platform with V3 chemistry (Illumina). We used the DADA2 v.1.24 pipeline56 in R to trim, quality-filter denoise and dereplicate the sequences, for generation of ASV tables and to assign taxonomies. The UNITE dynamic database, released on 2âFebruary 2019, and SILVA SSU r132, released in March 2018, were used for fungal and bacterial taxonomic assignment, respectively. Median read numbers were 32,200 (interquartile range (IQR) 25,631â38,508) and 38,410 (IQR 31,831044,246) for 16S and ITS, respectively. Two replicate sites from Spain (sites 1 and 3; nâ=â42) plus two further individual samples were excluded from analyses due to low DNA yield and poor recovery of reads, particularly for the prokaryotic amplicons, leaving 574âsamples in total.
To determine the functional gene composition of our initial samples and our microcosms at S1 and S4, the whole metagenome was sequenced by the Centre of Genomic Research (28âinitial samples (ES1 and ES3 excluded)â+â28 replicate sitesâÃâ2 sampling timesâÃâ5 treatmentsâ=â308 samples). The Illumina unamplified fragment library was prepared using the TruSeq PCR-free kit (350-base-pair (bp) inserts) and shotgun sequenced in four lanes of the NovaSeq platform using S4 chemistry (2âÃâ150âbp paired-end). Illumina adaptor sequences were detected and removed with Cutadapt (v.1.2.1), before trimming with Sickleâ1.200 using a minimum window quality score of 20 and with exclusion of reads shorter than 15âbp. Forward reads were then functionally annotated using DIAMOND BLASTX (v.0.8)57 using scripts and procedures from the SAMSA2 pipeline (v.2) and the included SAMSA formatted SEED subsystems database58. Median read number for metagenomes was 6,398,524 (IQR 4,704,883â8,216,799).
Statistical analyses
All statistical analyses were performed in Râsoftware v.4.2 or above59, and most plots were generated using ggplot2 v.3.3 (ref. 60). Râpackages used in custom code (Code availability) to assist core analysis and plotting were: broom.mixedâ0.2 (ref. 61), carâ3.1 (ref. 62), cowplotâ1.1 (ref. 63), eulerrâ7.0 (ref. 64), fsâ1.6 (ref. 65), ggordiplotsâ0.4 (ref. 66). ggrepelâ0.9 (ref. 67), Hmiscâ5.1 (ref. 68), lmerTestâ3.1 (ref. 69), magrittrâ2.0 (ref. 70), pacmanâ0.5 (ref. 71), CARTOcolorsâ2.1.2 (ref. 72), pracmaâ2.4 (ref. 73), RColorBrewerâ1.1 (ref. 74), seqinrâ4.2 (ref. 75) and tidyverse:2.0 (ref. 76).
For visualization of the relative importance of the effect of treatment, country, site and sampling time on taxonomic and functional gene abundance data, as well as measures of soil functioning, we ran NMDS analysis using the function metaMDS in the vegan package77, BrayâCurtis dissimilarity, followed by partial RDA, to visualize the relationships between disturbance treatments and community composition, controlling for country and site effects (vegan Râfunction rda). To test the effect of inclusion of different numbers of countries, total variance explained by disturbance treatments was summed across all four RDA axes. To quantify treatment effects on taxonomic and functional gene abundance data, as well as measures of soil functioning, PERMANOVA was performed on all except the initial samples, testing the effects of disturbance, sampling time and their interactions with country and site. This was implemented using the adonis2 function in the vegan package on BrayâCurtis dissimilarities among Hellinger-transformed counts and 999âpermutations. All above assessments of beta-diversity used rarefied data (rrarefy function in vegan), standardized to the sample with the lowest numbers of reads (2,043âbacterial reads, 4,091âfungal reads and 496,126âfunctional reads).
Individual resistance and resilience
For identification of different response categories, we fitted linear mixed-effects models to each ASV using the lme function in the nlme Râpackage v.3.1 (ref. 78). These models fitted fixed effects of disturbance treatment, sampling time (in days from S1) and their interaction on ASV relative abundance. Random effects on the intercept were included for country and site within country. Differences in variance (heteroscedasticity) were accounted for with sampling depth (accounting for both additive and proportional error, using the varConstProp function also in the nlme package), and with each of the sampling points (S1â4). In each case, the sign and significance of difference from zero (Pââ¤â0.05 or not by Wald test) were recorded for treatment effects (differences in relative abundance from control at S1) and interaction terms (differences in slope of relative abundance over time from control). These were used to classify the ASV response to each disturbance in terms of resistance (either significantly positively or negatively impacted or resistant, with no significant impact at S1) and resilience (either resilientâsignificant change in relative abundance over time in the opposite direction to the impact, or divergingâsignificant change over time in the same direction as the impact; or stableâno significant change over time).
Phylogenetic clustering of these strategies was determined by constructing a tree of the 500âmost abundant 16S and ITS ASVs, each aligned using kmer and secondary structure-guided alignment with AlignSeqs from the DECIPHER Râpackage v.2.24 (ref. 79). Trees were constructed using FastTree v.2.1 (ref. 80), with pseudocounts and a generalized time-reversible (gtr) model and rooted at the split between archaea and bacteria for both 16S sequences and fungi at the split between an outgroup comprising ASVs identified as belonging to the basal-branching phylum Chytridiomycota81 and the other taxa. For each disturbance, a phylogenetic least-squares model of both impact of disturbance and change over time (that is, the treatment and treatment:time effects for each ASV from the mixed-effects models described above) was fitted using the caper v.1.0 package in R82. This approach estimates a value and confidence interval for phylogenetic signal in terms of Pagelâs lambda83.
For estimation of the resistance and resilience of functional gene categories, the arcsine square-root-transformed relative abundances of functional genes at each level of the SEED Subsystems classification84 were first calculated. Mixed-effects models were fitted to these values with the lme4 v.1 package85. This model used treatment and sampling time as fixed effects, with site nested in country and all lower SEED Subsystems levels nested (level2/level3/level4) as random effects on the intercept. In addition, observation-level random effects (that is, one level for a particular read-count in a particular sample, for a particular gene in the SEED category being modelled) were fitted for each country, to allow for different levels of variance among countries (that is, heteroscedasticity). Estimated resistance and resilience effects were then extracted from the model in the same way as for the ASV models above (treatment and treatment:time interactions, respectively). Pâvalues were obtained for the significance of these treatment contrasts, accounting for multiple comparisons with the glht function in the multcomp v.1.4 package in R86, using Dunnettâs test and explicit planned contrasts for treatment and treatment:time, respectively.
Growth rates
We quantified bacterial growth across whole communities, using the ratio of bacterial origins and bacterial termini. Because bacterial chromosomes replicate from a single origin to a terminus, actively growing bacterial populations have, on average, a gradient in copy number increasing from terminus to origin87, the basis of tools used to identify the growth rates of particular microbes from metagenomic data88,89,90. We counted matches to dnaA protein sequences, the replication initiator protein as a marker of bacterial origins, where it is consistently found91, and to the dif DNA sequence, a broadly conserved 28âbp binding site for the chromosome dimer resolution machinery and a marker of bacterial termi92, markers specifically validated in a community context89. For each sample, we matched one of the paired-end metagenomic read files (R1) to databases of 39,401âdnaA sequences taken from RefSeq93 releaseâ215, and to a set of 578âdif sequences92. For identification of dnaA sequences, we used DIAMOND v.2.0 (ref. 57) in translated (blastx) mode set to âvery sensitiveâ, counting every sequence in which at least one match had an e-value of 0.001 or less as a hit. For identification of dif sequences, we used nucleotide BLAST v.2.13 (blastn94) with the blastn-short task settings, counting every sequence in which at least one match had an e-value of 0.01 or less as a hit. Counts of origin and terminus hits were converted to estimates of relative bacterial growth by taking the difference between log2 of the ratio of origin hits to terminus hits in each treatment sample and the equivalent value in the unperturbed control sample. These growth rate estimates were analysed using a mixed-effects model with fixed effects of treatment, sampling time, BrayâCurtis dissimilarities from control of the bacterial and fungal communities and all possible interactions among these effects. Random effects on the intercept of site nested within country were included, along with an effect of treatment on variance, using the lme function in R as above. This complete model was reduced to a minimal adequate model by stepwise removal of non-significant effects using the stepAIC function from the MASS package v.7.3 (ref. 95). This reduced model contained only main effects of treatment, sampling time, the distance of the fungal community from control and the interaction between that distance and treatment.
Growth capacity
Growth capacity was quantified using RasperGade16S v.0.0.1 (ref. 96) to estimate 16S copy number per cell for each prokaryotic ASV. This gave 1 as the most frequent copy number, 2 as the median copy number and a range of 1â18. The degree of confidence in that estimate (complete confidence, 1) was highly variable (median, 0.58; range, 2.6âÃâ10â4âââ1.00). These values were combined with ASV abundances to create a weighted average expected copy number for each sample, which was compared between treatment and control.
Community resistance and resilience
The resistance and resilience of microbial community structures (prokaryotes, fungi and metagenome, as opposed to individual ASVs or functional categories) were defined as the negative BrayâCurtis dissimilarity between disturbance and control at S1 and S4, respectively (that is, if the distance was high, the resistance/resilience was low)14,97. To test for relationships between initial soil, climatic and microbial properties and microbial community responses to extreme events (resistance and resilience), as well as between those initial properties and the initial relative abundance of each of the highest-level functional categories and between the community response and those initial functional categories, we used rank (Spearman) correlations across samples. We also used rank correlations between distance matrices measuring BrayâCurtis dissimilarity among the relative abundances of functional categories and the Euclidean distance among rank-transformed individual soil properties for each set of soil properties (enzymes: phosphatase, β-glucosidase, leucine-aminopeptidase and acetylesterase; MicroResp: water, alanine, cellulose, citric acid, fructose, glucose, glycine and malic acid; soil C and N availability: DOC, DON, nitrate and ammonium; gas fluxes: CO2, CH4 and N2O) at either the S1 or S4 time point. Pâvalues were calculated using 999âpermutations in the mantel() function in the vegan package. Heatmaps were clustered where necessary using complete-linkage hierarchical clustering on the values of the correlations.
Predictive model
An explicit predictive model of resistance and resilience (negative log-transformed BrayâCurtis dissimilarity of metagenome gene abundances in treatment from their control at S1 and S4, respectively) was fitted based on the 20âinitial soil and environmental properties (four temperature variables, four precipitation variables, eight carbon and nitrogen variables, three water-holding variables and pH), along with treatment and sampling time. These explanatory variables were each standardized to a mean of zero, standard deviation of 1 and used as explanatory variables in a random forests model98. That regression forest was fitted using the randomforest Râpackage (v.4.7)99 with 10,000âtrees and a default mtry parameter (one-third of explanatory variables randomly sampled at each split). A modified grouped cross-validation of this model was carried out, in which the data were split into training and test sets and with each training set containing data from two replicate sites in each country; the test set contained data from the third site. There are 19,683âunique ways of doing this split (after exclusions, see above). A random forest model, as above, was fitted to each of these training sets and used to make predictions for each respective test set. The proportion of variance explained (coefficient of determination) was calculated for each test set using the random forest function. An alternative grouped cross-validation was carried out where training sets comprised the data from six countries, and test sets comprised those from the remaining four countries (these proportions were chosen because they enable an estimation of error for each country and are close to the threefold cross-validation proportions used above). There are 210âunique ways of doing this split, with models fitted and coefficients of determination calculated for each one as above. For both cross-validations, Pearson correlations were calculated between observed values and the mean value predicted across cross-validation models for each site within each perturbation. Partial dependence plots, which visualize the relationship between a variable of interest and the response, also accounting for the average effect of other predictors100, were created using the pdp Râpackage (v.0.8)101.
Ethics and inclusion statement
This project was planned and executed in close collaboration with local partners. Sampling adhered to local laws, and permission was obtained from landowners. All local partners have been included as coauthors.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.