Fly stocks
All experiments used mated female flies unless otherwise stated. Flies were reared on standard cornmeal molasses food at 25â°C and 50% humidity. For optogenetic activation experiments, flies were reared in the dark on standard food supplemented with retinal (Sigma-Aldrich) unless otherwise specified, 0.2âmM all trans-retinal before eclosion and 0.4âmM all trans-retinal after eclosion. Hemidriver lines were created using gateway cloning as previously described61. Stable split GAL4 lines used in this study were constructed as described previously61 and hemidrivers used are described in Supplementary Table 1. iLexA stands for improved LexA, in which additional activating domains were added the transcription activator to enhance expression62. For the aIPg-iLexA line, the pBPnlsLexA::p65::GADUw vector was used for cloning as described previously48). Original confocal image data of GAL4 lines are available at https://www.janelia.org/split-GAL4. Genotypes used in each figure are listed in Supplementary Tables 1, 3â5, 7, 9â11, 13, 15, 17, 18, 20â22, 24, 26â30, 32â34, 36 and 37.
Thermogenetic and optogenetic activation behavioural experiments
Groups of 5â8 group-housed mated female flies (7â10 days after eclosion) were video recorded at 60% relative humidity in a 53.3âmmâÃâ3.5âmm circular arena. This assay and automated analysis pipeline was used as described63. All non-thermogenetic (TrpA) experiments were performed at 24â°C, while thermogenetic experiments were performed at 22â°C for non-activating controls and 31â°C for activation experiments. All tests were conducted under visible light conditions at zeitgeber time 0 to zeitgeber time 4 unless otherwise stated. Flies were loaded into the arena using an aspirator. For activation of neurons expressing Chrimson, the arena was illuminated as specified in the figure legends using constant uniform illumination with 660ânm LEDs. For inactivation of neurons expressing GtACR, we used constant uniform illumination with 525ânm LEDs. All of the trials were performed under white-light illumination from above. Videos were recorded from above using a camera (USB 3.1 Blackfly S, Monochrome Camera; Point Grey) with an 800ânm long-pass filter (B+W filter; Schneider Optics) at 170 frames per second and 1,024âÃâ1,024 pixel resolution.
Behavioural classification and analysis
For each trial, flies were acclimatized to the arena for 30âs before the delivery of six sets of constant stimuli each lasting 30âs with 30âs in between each stimulus. For all experiments, only the lowest stimulus intensity in which an effect was found is depicted and was analysed. Unless otherwise stated, the pre-stimulus average was calculated from the three periods before the stimulus periods used for analysis. In Fig. 3f, the previous stimuli appeared to alter behaviour during successive stimulus-off periods; therefore, only the first 15âs of the first pre-stimulus period was used for comparison. The videos were processed using the automated pipeline described previously63. In brief, flies were tracked using Caltech FlyTracker followed by automated classification of behaviour with JAABA classifiers64. Novel classifiers for touch and aggression were created based on previous definitions46,65. We validated the performance of these classifiers against manually labelled ground-truth data using videos that were not part of the training dataset (framewise performance is shown in Supplementary Table 2). As aggression is inherently the interaction of more than one fly, we cannot disassociate the behaviour of one fly from that of the others. Aggression is therefore computed on a per-arena basis rather than a per-fly basis. The percentage aggression is the sum of the aggression scores for all the trajectories in which flies were performing aggression divided by the total number of flies in the arena times 100. For figures displaying behavioural time courses, the mean of 0.35âs (60-frame) bins is shown. The angle occluded by nearest fly was calculated using the anglesub perframe feature65 and was computed by finding the two lines tangent to the fit ellipse that intersect at the nose-point of this fly, and measuring the angle between them.
For dyadic courtship assays (nâ=â13), the courtship start frame was manually identified based on first-instance inter-fly distance of <3âmm and fixation angle of <|20°| lasting for >1âmin. Courtship end was defined as first copulation frame or end of video acquisition (30âmin).
For calculation of visual features experienced during aggressive and courtship interactions, the angular position, velocity, height and width were calculated on a frame-by-frame basis using a custom MATLAB (MathWorks) script whereby, for each frame, the coordinates and orientations of subject fly and nearest conspecific, or target fly, were translated and rotated such that the subject was situated at the origin facing zero degrees. In this new basis, the target flyâs angular position (θ) and velocity (Ï) with respect to the subject flyâs visual field were calculated as \(\theta ={\tan }^{-1}\left(\frac{y}{x}\right)\) and \(\varphi =\frac{{\rm{d}}\theta }{{\rm{d}}t}\,\), respectively. To approximate the angular size and expansion of the target fly in the subjectâs visual field, an ellipse was fit to the major and minor axes of the target fly in this new basis. The target flyâs angular width was approximated as the length of the cross-section of this ellipse that lies perpendicular to the Euclidean line between the anterior-most point of the subject fly and target fly centroid. Thus, for each frame, the equation for the target flyâs angular width, w, is \(w=2\left({\tan }^{-1}\frac{R}{d}\right)\), where R is half the real cross-sectional length of the ellipse (in mm) and d is the Euclidean distance (in mm) between the anterior-most point of the subject fly and target fly centroid. The target flyâs angular height was approximated in a similar manner; however the target flyâs real height was fixed at 1âmm (which a reasonable estimation of a female flyâs height), so the equation for angular height, h, was simply \(h=2\left({\tan }^{-1}\frac{0.5\,{\rm{mm}}}{d}\right)\), where d is the Euclidean distance (in mm) between the anterior-most point of the subject fly and target fly centroid. Female aggression frames were defined using the JAABA aggression classifier and calculated from 79 trajectories.
Fly preparation for calcium imaging
For Fig. 3a and Extended Data Fig. 2aâd, experiments were performed similarly to those described previously24 with a similar preparation to that described in a previous study66. Notably, the flyâs head was positioned and glued to the fly holder such that the eyeâs equator faced the middle of the visual projection screen. The proboscis remained intact but was glued in position, and a dissection needle was used to remove the cuticle and sever muscle 16.
For Fig. 6c, experiments were performed similarly to as described previously11. In brief, the flies were anaesthetized on CO2 and tethered to a custom-milled plate. The flies were held in place by a string across the neck and fixed to the holder by both eyes and the back of the thorax using UV-curable glue. The proboscis was also glued to the mouthparts to minimize brain motion. Flies were left to recover in a warm, humidified chamber (25â°C, 50â70% humidity) in the dark for 1â4âh. The cuticle was subsequently dissected from the top of the head and flies were transferred to an air-supported foam ball.
Two-photon calcium imaging
In Fig. 3a and Extended Data Fig. 2aâd, calcium imaging experiments were performed with male or female flies (LC10a and LC9 experiments respectively) 5â10âdays after eclosion, maintained under standard conditions (21.8â°C, 55% humidity, 16âhâ8âh lightâdark, standard cornmeal/molasses food). A full list of fly genotypes used in the calcium imaging experiments is provided in the Supplementary Information. The imaging setup is identical to the previously described two-photon microscope (Thorlabs) setup24. In brief, we used a Ti:Sapphire femtosecond laser (Spectra-Physics Mai Tai eHP DS) tuned to 920ânm and delivering <20âmW power at the sample. Fluorescence signals were collected using a Ã16 water-immersion objective (Nikon CFI75, NA 0.8) with a band-pass filter (Semrock 503/40ânm) in front of the photomultiplier tube (Hamamatsu GaAsP H10770PB-40 SEL). Oxygenated saline was circulated throughout. Imaging volumes were acquired at 5.6âHz or higher. Visual stimuli were delivered to the flyâs right eye and all imaging was from the right side of the brain. The stimuli were presented on a screen that subtended approximately 90° by 90° of the flyâs field of view with a green (532ânm) projector setup as previously described24.
In Fig. 6c, calcium imaging experiments were performed with TuTuA_1 and TuTuA_2 male flies 3â7 days after eclosion, maintained under standard conditions (25â°C, 65% humidity, 12âhâ12âh lightâdark, standard Würzburg food). The imaging preparation for tethered courtship was identical to that previously described11. In brief, male flies rested and walked on a small 6.35âmm diameter ball, which was shaped from foam and manually painted with uneven black spots using a Sharpie. The foam ball was held by a custom-milled aluminium base and floated by air supplied at ~0.8âlâminâ1 such that the ball could move smoothly. The ball was illuminated by infrared LED flood lights, and imaged with a Point Grey FLIR Firefly camera using a mirror. The ball was surrounded by a 270° conical screen with a large diameter of ~220âmm, a small diameter of ~40âmm and a height of ~60âmm. As males walked on the foam ball, all three rotational axes of the ball were read out by the FicTrac2.0 software67 at 60âHz in real-time. The visual stimulus was projected around the male from a DLP 3010 Light Control Evaluation Module (Texas Instruments) through a first-surface mirror below the fly. The red and green LEDs in the projector were turned off, leaving only the blue LEDs to minimize interference with GCaMP emissions.
Visual stimuli were generated in the MATLAB-based ViRMEn software68 and projected onto the screen using custom perspective transformation functions. The net visual refresh rate of the visual stimulus ranged from 47.6âHz to 58.9âHz. Each trial was initiated by the presentation of a stationary visual target for 60âs to examine the animalâs baseline locomotion, after which the visual target began to oscillate. The visual target oscillated in a 107° arc around the animal with a constant angular velocity of approximately 75°âsâ1, but the angular size of the dot was continuously altered to mimic the dynamics of a natural female during courtship. The angular size was altered by changing the distance between the male and the target in the ViRMEn world. The distance between the male and the target was taken from the inter-fly distance in a courting pair over the course of two minutes of courtship and, at each frame, the angular position of the target was scaled by this inter-fly distance to give rise to a more dynamic female path. Angular sizes ranged between around 8 and 50°, with the average size being 22.5°. Each stimulus frame was therefore unique for 2âmin of time, and subsequently repeated until the end of the trial when it intersected its original position. Each trial lasted 10âmin.
Male imaging experiments were performed using the Ultima Investigator or Ultima Investigator Plus two-photon laser-scanning microscope (Bruker Nanosystems) with a Chameleon Ultra II Ti:Sapphire laser. All of the samples were excited at a wavelength of 920ânm, and emitted fluorescence was detected with a GaAsP photodiode detector (Hamamatsu). All images were acquired using a Ã40 Olympus water-immersion objective with 0.8âNA. All images were collected using PrairieView Software (v.5.5 or 5.7) at a resolution of 512âpxâÃâ512âpx.
Courtship and running was classified based on the fidelity and vigour of a maleâs pursuit of the visual target, as described previously11.
In Extended Data Figs. 5b and 7aâe, ex vivo calcium imaging experiments were performed similarly to those described previously69. In brief, flies were reared at 25â°C on cornmeal medium supplemented with retinal (0.2âmM) that was shielded from light. All experiments were performed on female flies, 3â5 days after eclosion. Brains were dissected in a saline bath (103âmM NaCl, 3âmM KCl, 2âmM CaCl2, 4âmM MgCl2, 26âmM NaHCO3, 1âmM NaH2PO4, 8âmM trehalose, 10âmM glucose, 5âmM TES, bubbled with 95% O2/5% CO2). After dissection, the brain was positioned anterior side up on a coverslip in a Sylgard dish submerged in 2âml saline at 20â°C. The sample was imaged with a resonant scanning 2-photon microscope with near-infrared excitation (920ânm, Spectra-Physics, INSIGHT DS DUAL) and a Ã25 objective (Nikon MRD77225 25XW). The microscope was controlled using ScanImage 2017 (Vidrio Technologies). Volumes were acquired with a 230âμmâÃâ230âμm field of view at 512âÃâ512âpx resolution at 2 âμm steps over 42 slices, at approximately 1âHz. The excitation power for Ca2+ imaging measurement was 15âmW. On the emission side, the primary dichroic was Di02-R635 (Semrock), the detection arm dichroic was 565DCXR (Chroma), and the emission filters were FF03-525/50 and FF01-625/90 (Semrock). During photostimulation, the light-gated ion channel Chrimson was activated with a 660ânm LED (M660L3 Thorlabs) coupled to a digital micromirror device (Texas Instruments DLPC300 Light Crafter) and combined with the imaging path with a FF757-DiO1 dichroic (Semrock). Photostimulation occurred at 10âHz over two periods with a duration of 14âs at 0.037âmWâmmâ2 intensity interspersed by a 2âs pause. After responses to the photostimulation, the laser power was increased to take two-colour high-resolution images containing fluorescence from both the red and green channels. Using custom Python scripts, regions of interest (ROIs) corresponding to cell compartments were identified in the high-resolution images. These ROIs were then applied to the time-series images to measure intensity changes in response to the photostimulation. Fluorescence in a background ROI, which contained no endogenous fluorescence, was subtracted from the cell compartment ROIs. In the ÎF/F calculations, baseline fluorescence is the mean fluorescence over a 10âs time period before stimulation started. The ÎF is the fluorescence minus the baseline. The ÎF is then divided by the baseline to normalize the signal (ÎF/F). Outlier samples with very low intensities or those of which the intensity randomly fluctuated were excluded from the analysis.
Electrophysiology
Whole-cell patch-clamp recordings were obtained from freshly isolated brains of 3â5 day old flies. The brain was continuously perfused with oxygenated (95% O2/5% CO2) extracellular saline containing 103âmM NaCl, 3âmM KCl, 1.5âmM CaCl2·2H2O, 4âmM MgCl2·6H2O, 1âmM NaH2PO4·H2O, 26âmM NaHCO3, 5âmM TES, 10âmM glucose and 10âmM trehalose·2H2O. Osmolarity was 275âmOsm and the pH was 7.3. Recording electrodes were pulled from thick-walled glass pipette (1.5âmm/0.86âmm) using the P-97 puller (Sutter Instruments) and fire polished using MF 830 (Narishige) to achieve resistances of 10â12âMΩ. Intracellular saline contained 137âmM KAsp, 10âmM HEPES, 1.1âmM EGTA, 0.1âmM CaCl2·2H2O, 4âmM MgATP, 0.5âmM NaGTP. Osmolarity was 260â265âmOsm and the pH was adjusted to 7.3 with KOH. Biocytin was added to intracellular solution at 0.5% for post hoc morphological confirmation.
The brain was visualized by an IR-sensitive CCD camera (ThorLabs 1501M) with an 850ânm LED (ThorLabs M850F2). GFP-labelled cell body was visualized with 460ânm LED (Sutter Instruments). Images were acquired using Micro-Manager with automatic contrast adjustment. Recordings were obtained from cell bodies under a 60Ã water-immersion objective (Olympus).
Current-clamp recordings were sampled at 20âkHz, low-pass filtered at 10âkHz using the Digidata 1550B system, Multiclamp 700B system and Clampex v.11.2 software (Molecular Devices). Recordings were made at a membrane potential of â50 mV to â65 mV, with small (5â30âpA) hyperpolarizing current injections as needed, and not corrected for liquid junction potentials.
Chrimson was activated by 630ânm LED at 0.4âmWâcmâ2. The stimulation duration was set at minimal value that is sufficient to induce reliable responses from target neurons. After the electrophysiology recording, the whole brain was fixed in 4% paraformaldehyde in 0.1âM PBS until further staining. After rinsing in PBS, the brain was incubated in Streptavidin Alexa Fluor 647 (1:200) in PBS-T overnight at room temperature. The preparations were then rinsed, dehydrated and mounted with DPX. The confocal images were captured on the LSM 980 microscope (Zeiss), with 639ânm excitation wavelength.
The electrophysiological recordings were analysed using pClamp (Clampfit v.11.3). The instantaneous action potential frequency was calculated for about 1âmin in each cell. The action potential amplitude was averaged from 20â30 individual events in each cell, and measured as the difference between the threshold and peak.
Immunohistochemistry and imaging
All experiments were performed as described previously46,70,71,72,73,74. Additional details of the imaging pipeline used are available online (https://data.janelia.org/pipeline).
Connectomics analyses
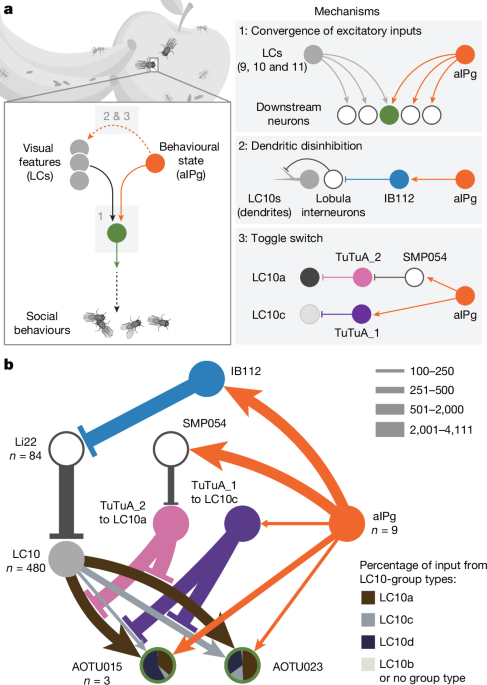
Our analyses are based on the hemibrain dataset25 (v.1.2.1) as queried using the neuPrint interface (https://neuprint.janelia.org/) unless otherwise noted. The unique identifier (bodyID number in the hemibrain v.1.2.1 database) for neurons is shown in the figures, and a complete list of synaptic connections used to construct our circuit diagrams can be found in neuPrint. LC10s in the hemibrain dataset were assigned to candidate types based on connectivity and morphology differences. As the hemibrain did not include the entire lobula, we also performed analyses in the recently completed and fully annotated male optic lobe connectome53 (neuprint release: optic-lobe: v.1.0) and the Flywire54 (v.783) analysis of the FAFB datasets75.
Synaptic connections organized by neuron pair and neuropil for FlyWire or optic lobe were obtained from Zenodo76 and neuPrint (using neuprint-python fetch_adjacencies with the default settings), respectively. The connection tables obtained in this way do not include synapses with small, untyped EM bodies as, in most cases, unidentified fragments of larger reconstructed cells are included in the tables and we did not include such synapses when calculating relative connection strengths. Including these additional synapses in calculations of relative connections strength (as done for example in the neuPrint web interface) results in lower numbers that most likely underestimate the relative contribution of one cell type to the inputs or outputs of another type. Although we did observe highly similar relative connection strengths for many cell type pairs, we also note that there were methodological differences between the FlyWire/FAFB and optic lobe datasets that may result in significant non-biological differences between synapse counts.
Neurotransmitters for TuTuA_1 and IB112 were determined by EASI-FISH77 using lines SS77547 for TuTuA_1 and SS81529 for IB112. Neurotransmitters for aIPg46 and LC10s78 were previously reported. Other reported neurotransmitters are based on computational predictions53,79.
Cell type annotations for FlyWire neurons59,60 were as downloaded from Codex on 24 April 2024 from https://codex.flywire.ai/api/download?data_product=visual_neuron_types&data_version=783. Matches of cell type names between the optic lobe and FlyWire datasets were as published53. FlyWire data to evaluate connections to descending interneurons described in Extended Data Fig. 10 were accessed through Codex (https://codex.flywire.ai) and evaluated using Flywire.
Statistics
No statistical methods were used to predetermine sample size. Sample size was based on previous literature in the field and is provided in the Supplementary Information (Supplementary Tables 6, 8, 12, 14, 16, 19, 23, 25, 31 and 35). Experimenters were not blinded in most conditions as all data acquisition and analysis were automated. Experiments were not randomized as most controls were performed within animals. Biological replicates completed at separate times using different parental crosses were performed for each of the behavioural experiments. Behavioural data are representative of at least two independent biological repeats. For figures in which the behavioural data over the course of a trial are shown, a yellow or red bar indicates the stimulus period, the mean is represented as a solid line and shaded error bars represent the variation between experiments.
For each experiment, the experimental and control flies were collected, treated and tested at the same time. A Wilcoxon matched-pairs signed-rank test (two-tailed) was used for statistical analysis of optogenetic experiments when examining effects within the same group. The results of two-way analysis of variance followed by Tukeyâs test (three or more groups) or uncorrected Fisherâs least significant difference test (two groups) for multiple comparisons for each optogenetic experiment are also reported in the Supplementary Information. For analysis among two groups, MannâWhitney U-tests (two-tailed) were used, while KruskalâWallis tests with Dunnâs multiple-comparisons post hoc analysis were used to compare across multiple groups. All statistical analysis was performed using Prism (GraphPad, v.10). P values are indicated by asterisks. The exact P values for each figure are provided in the Supplementary Information.
For bar plots, all datapoints are shown to indicate the range and the top edge of the bar represents the mean. The box plots show the median and interquartile range. The lower and upper whiskers represent 1.5âÃâ interquartile range of the lower and upper quartiles, respectively; the boxes indicate lower quartile, median and upper quartile, from bottom to top. When all points are shown, the whiskers represent the range and the boxes indicate the lower quartile, median and upper quartile, from bottom to top. In the violin plots, the lower and upper quartiles are indicated by dotted light grey lines, and the median is indicated by a solid light grey line. Shaded error bars on graphs represent the mean ± s.e.m.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.