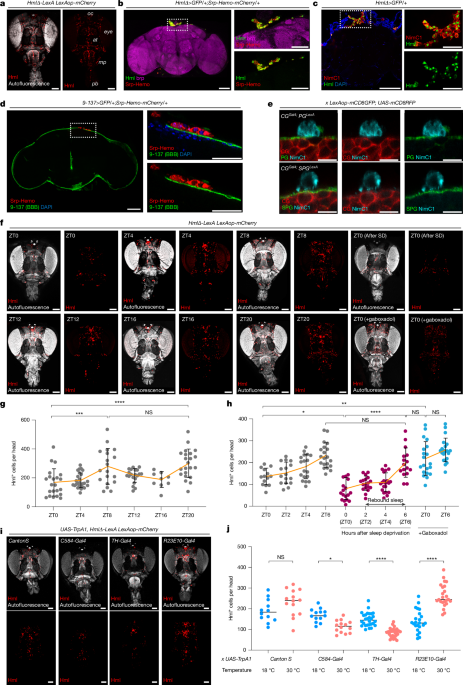
Drosophila strains and fly husbandry
All flies for experiments were maintained at 25 °C in a 12 h–12 h light–dark cycle, except for the temperature-sensitive Gal80 experiment in which Gal80 flies were kept at 18 °C until the experiment and activated Gal4 at 30 °C for 2 days. For the gene switch experiment, 500 μM mifepristone (RU486, sigma, catalogue no. M8046) was added to sucrose/agar food. For the RU486 control food, the same amount of 80% ethanol was added to sucrose/agar food. The following Drosophila stocks were used in this study: CantonS (Sehgal laboratory stock), HmlΔ-LexA LexAop-mCherry (J. Shim), HmlΔ-Gal4 UAS-EGFP (BL30139, BL30140), Srp-Hemo-mCherry (BL78358, BL78359, BL78362, BL78363), 9-137-Gal4 (Sehgal laboratory stock), NP2222-Gal4 (Sehgal laboratory stock), C584-Gal4 (Sehgal laboratory stock), TH-Gal4 (BL8848), R23E10-Gal4 (BL49032), R85G01-LexA (BL54285), R54C07-LexA (BL61562), Srp-Gal4 (L. Waltzer), Srp-Hemo-Gal4DBD (I. Evans), Srp-Hemo-Gal4AD (I. Evans), eater-dsRed (U. Banerjee), Ppn-Gal4 (BL77733), NP6293-Gal4 (Sehgal laboratory stock), moody-Gal4 (Sehgal laboratory stock), Repo-GeneSwitch (Sehgal laboratory stock), UAS-CD4::GRASP (BL58755), eater1 (BL68388), UAS-eater (BL36325), eater RNAi (BL25863, V4301), UAS-mCD8GFP (BL5137), UAS-mCD8::RFP, LexAop-mCD8::GFP (BL58754) UAS-LD::GFP (M. Welte), UAS-GFP::LSD2 (M. Welte), UAS-Sirt1 (BL44216), GLaz-GFSTF (BL60526), UAS-h.MEGF11(BL78460), MZ0709-Gal4 (M. Freeman), Alrm-Gal4 (M. Freeman), Repo-Gal4 (L. Griffith), Repo-LexA (M. Freeman), put RNAi (BL35195), Lsd-1 RNAi (V30884), NimC3 RNAi (V22920), GLaz RNAi (V4806), Lsd-2 RNAi (V40734), UAS-PvrDN (BL58431), Nplp2 RNAi (BL54041), NimC1 RNAi (BL25787), NimC4 RNAi (BL61866), NimB1 RNAi (BL55937), NimC2 RNAi (BL25960), Smox RNAi (BL41670), PGRP-LC RNAi (BL33383), drpr RNAi (BL67034), NimB4 RNAi (BL55963), Apoltp RNAi (BL 51937), NimA RNAi (V104204), Col4a1 RNAi (BL44520), NimB2 RNAi (BL62289), NimB5 RNAi (V15758), babo RNAi (BL25933), Karl RNAi (V9446), LpR2 RNAi (BL31150), apolpp RNAi (BL28946), Acsl RNAi (V3222), LpR1 RNAi (BL27249), crq RNAi (BL40831), NimB3 RNAi (V330502).
Sleep recording
For the fly sleep recording, mated 5- to 7-day-old flies were loaded into glass tubes containing 5% sucrose with 2% agarose. At least 2 days after loading into the monitors, sleep was analysed for 3 days. Single beam monitors were used for RNAi screening and the sleep deprivation experiment, but other sleep recordings were performed with multibeam monitors. Sleep was defined as failure of the fly to cross the red beam in the monitor for 5 or more minutes, analysis of data was performed with an in-house built code as described previously56.
Sleep deprivation
We used mechanical sleep deprivation. To achieve deprivation, flies in single beam monitors were fixed to a vortex machine and shaken randomly for 2 s every 20 s over a 12-h period (from ZT12–ZT24). To estimate rebound sleep, 6 h of sleep after sleep deprivation was compared with sleep on the pre-deprivation day at the same ZT time (ZT0–ZT6). Analysis of per cent sleep gain was as described previously57. In short, 6 h of daytime sleep on the day before deprivation was subtracted from the 6 h after deprivation (sleep gain). Similarly, sleep loss was calculated by subtracting sleep during deprivation to night-time sleep a day before the deprivation (sleep loss). Percentage of sleep gain was calculated by amount of sleep gain relative to amount of sleep loss.
Fly tissue clearing and haemocyte quantification
For visualizing circulating haemocytes, we optimized two different protocols58,59. Heads of female HmlΔ-LexA LexAop-mCheery flies were cut with a micro-scissor and fixed with 4% paraformaldehyde for 4 h at room temperature with rotation. After fixation, heads were incubated with 100% methanol at 4 °C overnight. Methanol was removed the following day and heads were incubated with BABB solution (2:1 ratio of benzyl benzoate and benzyl alcohol) for at least 6 h. After removing the BABB solution, heads were mounted on glass slides with VECTASHIELD solution without 4′,6-diamidino-2-phenylindole (DAPI). Imaging was performed immediately after mounting and a 408-nm excitation laser was used for auto-fluorescent signals. For thermogenetic experiments where wake or sleep neuronal populations were manipulated, flies were maintained at 18 °C until TrpA1 was activated for 1 day at 30 °C. Following this, they were processed as above. For haemocyte counts, we used the three-dimensional (3D) object counter in ImageJ software.
Immunohistochemistry
Brains were dissected from female flies, fixed in a 4% paraformaldehyde solution and washed three times using 0.4% PBS TritonX-100. After three washes, samples were blocked using 10% normal goat serum for 30 min at 25 °C. Samples were incubated with the desired primary antibodies overnight at 4 °C and then washed three times using 0.4% PBS TritonX-100 (PBST). Samples were incubated with secondary antibodies (Life Tech, catalogue nos. A32723, A32740, A32742, A32731 and A21236) diluted 1:250 for 2 h. Samples were then washed three times with 0.4% PBST. After washing, samples were rinsed and kept in VECTASHIELD until they were mounted on glass slides. For haemocytes count, we did not remove air sacs to maximize the number of haemocytes. The following primary antibodies were used: α-NimC1 (a gift from I. Ando; 1:100), α-brp (Developmental Studies Hybridoma Bank (DSHB), catalogue no. nc82; 1:100), α-Repo (DSHB, catalogue no. 8D12; 1:100), α-cleaved dcp1 (Cell Signaling, catalogue no. 9578S; 1:100), Oil-Red O (Sigma, catalogue no. O9755) and BODIPY 493/503 (Fisher, catalogue no. D3922, 1:1,000). Images were obtained with a Leica Stellaris STED confocal microscope. For haemocyte counts, we used the 3D object counter in ImageJ software.
Staining of LDs using Oil-Red O
Brains from female flies were dissected and fixed as for immunohistochemistry. After fixation, brains were washed three times with 0.4% PBS TritonX-100 and kept in 0.4% PBST overnight at 4 °C. If a sample needed primary antibody incubation, it was blocked with 10% normal goat serum in 0.4% PBS TritonX-100 for 30 min and then kept in diluted antibody with 0.4% PBST overnight at 4 °C. The following day, Oil-Red O solution (0.1 g per 20 ml of isopropanol; Sigma, catalogue no. O0625) was prepared in 0.4% PBST as a 2:3 ratio. If not stained with primary antibody, the sample was incubated with Oil-Red O solution for 10 min and washed with distilled water five times for 5 min each. If samples were stained with primary antibody, samples were washed and treated with secondary antibody and, after the secondary antibody, incubated with Oil-Red O solution. Finally, samples were rinsed with PBS and mounted in VECTASHIELD until they were mounted on glass slides. Images were obtained with the Leica Stellaris STED confocal microscope.
Quantification of LDs in the brain
To count LDs, images were analysed with a custom ImageJ macro. For each slice in the stack, the BioVoxxel toolbox was used to subtract background noise using the convoluted background subtraction method with a mean convolution filter of 3 radius. Once the background was subtracted, the image was duplicated, and one of the copies was converted into a mask that contained only the brain region. Inside the masked area, LDs were counted using the Analyze Particles tool, defining a particle size of 2–250 and circularity of 0.4–1.0. Reported results are the sum of particles in the whole stack. The macro is publicly available at https://github.com/CamiloGuevaraEsp/lipid_droplets.
Immunoprecipitation and western blot
In a 15-ml tube, at least 100 mixed-sex flies were collected for immunoprecipitation, or 20 flies for western blot. Flies were frozen on dry ice for 10 min. After freezing, flies were vortexed for 10–20 s three times to shake off heads. Flies were then poured into a sieve that allows passage only of heads. Fly heads were homogenized at 25 Hz for 2 min in a TissueLyser II (Qiagen) in 100 μl of lysis buffer for immunoprecipitation (250 mM Tris-HCl (pH 7.5), 250 mM NaCl, 1.5 M sucrose, 1% TritonX-100, protease inhibitor cocktail) or for western blot (RIPA buffer, Lifetech, catalogue no. 89901) with a 5-mm stainless steel bead (Qiagen, catalogue no. 69989) in round-bottom tubes (USA Scientific, catalogue no. 1620-2700). Homogenized samples were transferred to 1.7-ml microcentrifuge tubes and spun at 14,000 rpm for 10 min at 4 °C. Supernatants were used for the experiment. For immunoprecipitation, protein A/G magnetic agarose beads (Fisher, catalogue no. 78609) were used for antibody conjugation. Antibody conjugation to the bead or incubation of antibody with samples was performed in the cold room overnight. Samples were run in a 4–12% premade gel (Life Tech, catalogue no. NP0322). The following antibodies were used: anti-acetylated lysine-mouse (Life Tech, catalogue no. MA12021; 1:1,000), anti-acetylated lysine-rabbit (Cell Signaling, catalogue no. 9441S; 1:1,000), anti-DRP1 (a gift from L. Fisher; 1:1,000), anti-SRL (a gift from A. Duttaroy, 1:1,000), anti-α-tubulin (DSHB, catalogue no. 12G10; 1:1,000), anti-FLAG (Sigma, catalogue no. F3165; 1:2,000), anti-mouse-horseradish peroxidase (HRP) (Jackson Immuno, catalogue no. 715-035-151; 1:2,000), anti-rabbit-HRP (Jackson Immuno, catalogue no. 715-035-152; 1:2,000) and HRP signal was obtained with ECL substrate (Life Tech, catalogue no. 32209).
Flow cytometry
At least 100 HmlΔ-Gal4 UAS-EGFP fly heads were cut with a micro-scissor under the microscope and kept in a round-bottom tube with 1,000 μl of ice-cold Schneider’s medium (Life Tech, catalogue no. 21720024) until the cutting was finished. With a metal bead, fly heads were homogenized with a TissueLyser II (Qiagen) at 25 Hz for 2 min. Samples were centrifuged at 6,000 rpm, for 5 min at 4 °C. Supernatant was discarded and pellets were treated with 37 °C pre-warmed 100 μl of Collagenase Type C (100 mg ml−1, Worthington-Biochem, catalogue no. LS004140), 380 μl of PBS, 20 μl of Dispase II (100 mg ml−1, Sigma, catalogue no. D4693). Samples were incubated on a rotator for 15 min and pipetted with a 200 μl pipet every 5 min. Then, 200 μl of ice-cold PBS was added to the sample, which was transferred to a 1.7-ml microcentrifuge tube. Samples were spun at 6,000 rpm, for 5 min at 4 °C. The supernatant was discarded and pellets were resuspended in 500 μl of cold Schneider’s medium; 0.5 μl DAPI (1 mg ml−1) was added and, after a short vortexing, samples were spun at 6,000 rpm for 5 min at 4 °C. Again, supernatant was discarded, and samples were resuspended in 600 μl of Schneider’s medium. Debris or clumps were removed using a 40-μm strainer (Sigma, catalogue no. BAH136800040) and samples were transferred to a 5-ml fluorescence-activated cell sorting (FACS) sorting tube. We used an Aria FACS sorter (BD Biosciences) with a 100-μm nozzle. Usually, 100 fly heads yield approximately 400 GFP+ haemocytes after sorting. The detailed cell gating strategy is in Supplementary Fig. 1.
Haemocyte transfer
At least 30 larvae were dissected in Schneider’s medium (Life Tech, catalogue no. 21720024) and kept on ice during the dissection. Samples were spun at 6,000 rpm for 5 min at 4 °C. Haemocytes were resuspended with 100 μl of PBS for a cell density of 100–150 cells per microlitre. Using the microneedle, 2–3 μl of haemocytes were injected into the fly thorax. HmlΔ-LexA LexAop-mCherry haemocytes were used for validating haemocyte transfer.
In vitro haemocyte culture
Larvae were dissected in 15 μl of Schneider’s medium (Life Tech, catalogue no. 21720024). Haemocytes were transferred to Schneider’s medium containing Dil-labelled neutral, oxidized or acetylated LDL (1:100 dilution, Life Tech, catalogue nos. L3482, L34358, L3484) in the tube. Haemocytes were incubated on Teflon printed microscopic slides (Immune-Cell, catalogue no. 61-100-17) for 2 h in the cold room. After 2 h, haemocytes were fixed with 4% paraformaldehyde and washed three times with 0.4% PBST. Haemocytes were kept in VECTASHIELD with DAPI before the imaging; images were obtained with a Leica Stellaris STED confocal microscope.
NAM food feeding
Flies were raised on normal food for 5 days after eclosion and then transferred to sleep recording glass tubes that contain 5% sucrose and 2% agarose food with or without 10 mM nicotinamide (Sigma, catalogue no. 72345).
Gaboxadol feeding
Gaboxadol hydrochloride (Sigma, catalogue no. T101) was dissolved in the normal fly food at a 2 mM concentration. Flies were kept in this for 2 days.
Acetyl-CoA measurement
To measure acetyl-CoA in the fly head, ten female flies were collected in the 15-ml falcon tube and frozen on dry ice for 10 min. After freezing, flies were vortexed for 10–20 s three times to shake off heads. Flies were then poured into a sieve that allows passage only of heads. Ten fly heads were homogenized at 25 Hz for 2 min in a TissueLyser II (Qiagen) in extraction buffer from the acetyl-CoA colorimetric assay kit (Elabscience, catalogue no. E-BC-K652-M). After homogenization, all procedures followed the manufacturer’s instructions from the kit.
NAD+/NADH measurement
To measure NAD+/NADH in the fly head, ten male and female CantonS and eater1 mutant flies were collected around ZT6 in 1.5-ml tubes then flash-frozen on dry ice. After freezing, flies were vortexed, and ten heads were collected then placed in a 2-ml tube with metal beads and 1 ml of lysis buffer (1:1 PBS: extraction buffer (10% DTAB 0.2 M NaOH)). The heads were homogenized using a TissueLyser II (Qiagen) at 25 Hz for 2 min. The homogenized liquid was passed through homogenizer tubes (Invitrogen, catalogue no. 12183-026) in a centrifuge for 5 min at 15,000 rpm at 4 °C. To measure NAD+ and NADH individually, 200 μl of lysate was added to separate 1.5-ml tubes; 100 ml of 4 M HCl was added to the NAD+ tube, and both were heated for 15 min at 60 °C. After incubation at room temperature for 10 min, 100 μl of 0.5 M Trizma base buffer was added to the NAD+ tube, and 200 μl neutralization buffer (a 1:1 mixture of 0.5 M Trizma: 0.4 M HCl) was added to the NADH tube. NAD+ samples were diluted 1:5 and NADH samples were diluted 1:1 using dilution buffer. Samples and standards were prepared 1:1 with NAD+/NADH-Glo detection reagent (Promega, catalogue no. G9071) according to manufacturer’s instructions, seeded onto a 384-well plate, and measured using a BioTek Cytation 5 imaging reader and the accompanying Gen5 v.3.12 software. Individual data points are the mean of three technical replicates.
MitoSox staining
To stain the fly brain with MitoSox (Fisher, catalogue no. M36008), ten fly brains were dissected in Schneider’s medium and kept in medium until the dissection was finished. Brains were transferred to Schneider’s medium with MitoSox dye (final concentration 5 μM) and incubated for 10 min with rotation at room temperature. After the incubation, brains were washed with Schneider’s medium three times for 3 min with rotation. Brains were fixed with 4% paraformaldehyde for 2 min and rinsed with PBS. MitoSox signal was imaged immediately after mounting with VECTA SHIELD using a Leica Stellaris STED confocal microscope.
Memory experiment
The memory experiment was as published60. In brief, 100 flies, 3–6 days old and of mixed sex, from the same genotype, were starved in agarose for 18 h. On the following day, flies were trained at 25 °C in 70% humidity in a small chamber containing 1.5 M sucrose or water soaked Whatman paper with odours (1:200 ratio of 4-methylcyclohexanol or 1:80 ratio of 3-octanol in paraffin oil) for 2 min under red light. After training, flies were placed in the bidirectional choice apparatus, which has an odour in each end. To remove bias coming from the odour, appetitive training was performed reciprocally. For long-term memory, trained flies were kept again in the agarose food for 18 h and the memory experiment was performed without re-training.
Lifespan experiment
Three hundred age-matched female or male flies were collected for 5 days and transferred to vials of 30 flies each. Every 2 or 3 days, flies were flipped into new vials and the number of flies was counted.
Haemocyte sample preparation for mass spectrometry analysis
From the HmlΔ-Gal4 UAS-EGFP fly, haemocytes were sorted by flow cytometry and 4,000 GFP+ cells were sorted into a 1.7-ml microcentrifuge tube. GFP+ cells were centrifuged at 6,000 rpm for 5 min at 4 °C, and pellets were kept at −80 °C until lipid extraction. Lipid extraction from fly haemocytes was performed using a modified Bligh and Dyer method61. In brief, frozen cell pellets were thawed at room temperature for 10 min before the addition of 200 µl of ultrapure water to facilitate cell lysis. This was followed by the addition of 450 µl of methanol and 250 µl of HPLC-grade chloroform. During extraction, 5 µl of 1 µg ml−1 of an internal standard mixture comprised of equiSPLASH (Avanti Polar Lipids, catalogue no. 330731), fatty acid 16:0-d2 (Cayman Chemical) and carnitine 14:0-d3 (Cayman Chemical) was added to each sample. The samples were vortexed for 10 s to form a single-phase solution and incubated at 4 °C for 15 min. Subsequently, 250 µl of ultrapure water and 250 µl of chloroform were added, inducing phase separation. The samples were then centrifuged at 16,000g for 10 min. The organic phase, containing the extracted lipids, was transferred carefully to fresh tubes and evaporated using a vacuum concentrator to obtain dried lipid extracts.
The dried lipid extracts were reconstituted in 200 µl of a 3:1 methanol:chloroform (MeOH:CHCl3) mix containing 10 mM ammonium formate. Following reconstitution, all samples were analysed using MRM methods. An injection solvent containing 0.02 µg ml−1 EquiSPLASH (Avanti Polar Lipids, catalogue no. 330731) was used as a quality control sample to monitor peak stability over time.
Unbiased lipidomics using MRM-profiling
Lipidomic analyses were performed using an Agilent 6495C triple quadrupole mass spectrometer coupled to an Agilent 1290 Infinity II LC system with a G7167B autosampler. Samples were introduced into the Agilent Jet Stream (AJS) ion source by direct flow injection (no chromatographic separation). Mass spectrometry data were acquired for 3 min per injection. For each MRM scan, 8 μl of sample was injected. MRM methods were organized into 25 methods on the basis of the ten main lipid classes based on the LipidMaps database, spanning over a total of 3,000 individual lipid species. Triacylglycerols and diacylglycerols were divided into separate methods based on fatty acid neutral loss residues. All MRM data were processed using CLAW MRM36.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.