Plasmids
To visualize co-translational interactions, plasmids encoding human β-actin, a 3×Flag tag, and 24 MS2 stem-loop repeats were constructed. The β-actin-24×MS2 (MBSV5) sequence54 (Addgene #102718) was amplified and subcloned into pRetroQ-AcGFP-C1 (TaKaRa) via PCR amplification, replacing AcGFP1. Additionally, truncated versions, containing the first 20, 101, 168, 305 and 368 amino acids of β-actin, were generated by amplifying the respective fragment lengths and introducing a stop codon. The plasmid pUbC-NLS-ha-stdMCP-stdGFP55, which expresses the MS2 coat protein fused to GFP (MCP–GFP), was used without modification (Addgene #98916). For imaging post-translational interactions, plasmids encoding β-actin, a 10×SunTag, a 3×Flag tag, and Xbp1u+ were designed. A synthetic β-actin gene (Uniprot P60709) was ordered from Integrated DNA Technologies (IDT) and cloned into pHRdSV40-K560-24×GCN4_v444 (Addgene #72229) by replacing the kinesin-1 motor domain via PCR amplification. The 24×SunTag repeats were then reduced to 10, and the Xbp1u+ sequence45 was introduced using PCR. To generate a plasmid encoding β-actin with the G150P point mutation fused to 10×SunTag, 3×Flag tag and Xbp1u+ the Q5 Site-Directed Mutagenesis Kit (NEB) was used according to the manufacturer’s instructions. To estimate non-specific interaction of chaperone and SunTag alone, a plasmid encoding the 10×SunTag, 3×Flag tag and XBP1u+ sequence without β-actin was constructed by PCR amplification to excise the β-actin gene from the previously generated plasmid. The plasmid pHR-scFv-GCN4-sfGFP-GB1-NLS-dWPRE, which expresses an scFv against the SunTag epitope fused to sfGFP (scFv-sfGFP), was used without modification44 (Addgene #60906). To estimate the distance between mRNA and translating polypeptides, 24×MoonTag56 epitope was placed at the N-terminal end of β-actin-24×MS2 plasmid. Plasmids encoding MoonTag specific nanobody 2H10 were generously provided by M. Tanenbaum. To estimate coincidental collisions with TRiC, we analysed interactions between TRiC and the 500-kDa VCP (P97) complex, which has diffusion properties similar to PFD (Extended Data Fig. 6l). A plasmid encoding VCP with C-terminal HaloTag was provided by Z. Liu.
A plasmid expressing CCT4 with SNAP-tag was generated by inserting SNAP-tag in a loop region between S202 and V203, obtained from Integrated DNA Technologies and cloned into pCW57.1-MAT2A (Addgene #100521).
Cell lines
Human U-2 OS cells (ATCC, HTB-96) were grown in complete media with DMEM containing 4.5 g l−1 D-glucose (Gibco), 10% fetal bovine serum (Gibco), 1% penicillin/streptomycin (Gibco) and 1% non-essential amino acids (Gibco) at 37 °C and 5% CO2. Cells were either transfected with Lipofectamine 3000 (Thermo Fisher) or electroporated (Neon transfection system, Invitrogen) following the manufacturer’s instructions. Cells were regularly tested for mycoplasma contamination and no contamination was detected.
The monoclonal U-2 OS cell line expressing CCT4–HaloTag was generated using CRISPR–Cas9-mediated gene integration. A CCT4 specific single guide RNA (sgRNA) (ATCTCTAAGATCTACACTGG) was cloned into the pSpCas9(BB)-2A-Puro (PX459) vector, which encodes SpCas9 and puromycin resistance57 (Addgene #48139). U-2 OS cells were transfected with the Cas9–sgRNA vector and a donor encoding HaloTag using Lipofectamine 3000 following the manufacturer’s instructions. Forty-eight hours after transfection, cells were selected with 1.5 µg ml−1 puromycin (Gibco). Surviving cells were further labelled with a fluorescent HaloTag ligand (Promega) and sorted via fluorescence-activated cell sorting to obtain single clones. The same strategy was applied to generate PFD4–Halo and PFD4–SNAP stable cell lines using PFD4 specific sgRNA (TTCAGTTGTATGCAAAATTC).
The polyclonal U-2 OS cell line expressing RPL10A–SNAP-tag was generated using lentivirus transduction. Lentiviruses were produced and transfected as described58. Forty-eight hours after transfection, cells were selected with puromycin (Gibco) and sorted with a fluorescent SNAP-tag ligand (NEB).
The PFD3 knockdown stable cell line was generated using a PFD3 specific sgRNA (TAAAGTGTGTCTGTGGTTGG) using CRISPR–Cas9-mediated gene deletion. U-2 OS cells were transfected with the Cas9–sgRNA vector using Lipofectamine 3000 and selected with puromycin after 48 h transfection. Cells were further sorted to isolate single clones.
Biochemical assays
SDS–PAGE and immunoblotting
Protein samples were boiled in 2× SDS Sample Buffer (Sigma-Aldrich) at 95 °C for 5 min. Equal amounts of total protein were loaded on SDS polyacrylamide gels. Proteins were separated by electrophoresis on NuPAGE 4–12% Bis-Tris Protein Gels (Thermo Fisher Scientific) using NuPAGE MOPS SDS Running Buffer (Thermo Fisher Scientific) at 120 V. Proteins were transferred from polyacrylamide gels to polyvinylidene difluoride (PVDF) membranes (Roche) in transfer buffer (25 mM Tris-HCl pH 7.5, 190 mM glycine, 0.1% SDS, 20% methanol) at a constant voltage of 100 V for 1 h. Membranes were blocked in blocking buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% Tween-20, 5% low-fat milk) for 1 h at room temperature. Membranes were incubated with primary antibody in blocking buffer for 1 h at room temperature or overnight at 4 °C. Immunodetection was performed using: anti-β-actin (Abcam, ab8226, 1/1,000 dilution), anti-CCT4 (Merck, HPA029349, 1/1,000 dilution), anti-DYKDDDDK Tag (Cell Signal, 2368S, 1/1,000 dilution), anti-GAPDH (Merck, MAB374, 1/1,000 dilution), anti-HaloTag (Promega, G9211, 1/1,000 dilution), anti-prefoldin 3 (Santa Cruz, sc-390524, 1/200 dilution), anti-prefoldin 4 (Thermo Fisher, 16045-1-AP, 1/300 dilution), anti-RPL10A (Abcam, ab174318, 1/1,000 dilution) anti-RPL29 (Thermo Fisher, PA5-27545, 1/1,000 dilution), anti-GCN4 (SunTag) (Addgene, 218104-rAb, 1/1,000 dilution) and anti-SNAP-tag (NEB, P9310S, 1/1,000 dilution). Blots were then washed 3 times for 10 min with blocking buffer without low-fat milk at room temperature and incubated with secondary antibody, conjugated anti-mouse immunoglobulin G (IgG)–horseradish peroxidase (HRP) (Merck, A4416, 1/10,000 dilution) or anti rabbit IgG–HRP (Merck, A9169, 1/10,000 dilution), in blocking buffer for 1 h at room temperature. Blots were washed 3 times for 10 min and developed on Amersham Image Quant 800 control software 2.1.0.3. Images were analysed and quantified in Fiji.
HaloTag pulldown
U-2 OS cells were lysed in standard lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.5% IGEPAL CA-630, 20 U ml−1 apyrase, 1 mM PMSF, 2 mM DTT, cOmplete Protease Inhibitor, Benzonase) on an end-over-end rotor for 30 min at 4 °C. The lysate was cleared by centrifugation at 16,000g for 20 min at 4 °C. Protein concentration was determined by Bradford assay. Halo-Trap Magnetic Agarose beads (Chromotek) were equilibrated in ice cold lysis buffer and subsequently washed 2 times with ice cold washing buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 2 mM MgCl2, cOmplete Protease Inhibitor) containing 5% glycerol and 2 times in washing buffer without glycerol. Cleared lysate was added to equilibrated Halo-Trap beads and incubated for 3 h at 4 °C with vertical rotation. The beads were washed three times in washing buffer containing 5% glycerol and three times in washing buffer without glycerol. For immunoblotting the bound protein was eluted in 2× SDS Sample Buffer by boiling at 95 °C for 5 min. For mass spectrometry the bound protein was eluted and digested with Elution Buffer I (50 mM Tris-HCl pH 7.5, 2 M urea, 5 μg ml−1 trypsin, 1 mM TCEP-HCl) for 30 min at 400 rpm at 30 °C. The sample was then combined with Elution Buffer II (50 mM Tris-HCl pH 7.5, 2 M Urea, 5 mM iodoacetamide) and incubated overnight at 400 rpm at 32 °C. The reaction was stopped by adding 25% trifluoroacetic acid (TFA, Merk).
Kinetics of actin transit to TRiC reflecting TRiC-mediated folding
U-2 OS cells were electroporated with plasmid encoding either actin–SunTag–Xbp1u+ or actin(G150P)–SunTag–Xbp1u+. For cells expressing actin(G150P)–SunTag–Xbp1u+, 10 µM MG-132 was added 4 h prior to the puromycin chase. Twenty-four hours after transfection, cells were pre-treated with 300 µg ml−1 puromycin (Gibco) for 2.7 min. The time 0 (t = 0) sample was collected immediately after pre-treatment. The chase was performed at time points of 0.3 min, 1.3 min, 2.3 min, 3.3 min, 5.3 min and 9.3 min. Cells were washed two times with ice cold PBS supplemented with 100 µg ml−1 cycloheximide (CHX) and frozen in a dry ice 2-propanol bath. Cells were lysed with 500 µl standard lysis buffer. The lysate was cleared by centrifugation at 14,000g for 20 min at 4 °C. Protein concentration was determined by Bradford assay. Anti-HaloTag pull down and sample preparation for immunoblot were performed as mentioned above. Immunoblots were analysed and quantified using Fiji. The average intensity from three replicates was plotted. Folding half-time was determined by fitting the data in SigmaPlot 14.0 using an exponential decay function: f = y0 + a × e−b × x.
Cycloheximide chase
U-2 OS cells were electroporated with a plasmid encoding either actin–SunTag–Xbp1u+ or actin(G150P)–SunTag–Xbp1u+. Twenty-four hours after electroporation, cells were treated with 150 µg ml−1 CHX (Sigma-Aldrich) for 0.5 h, 0.75 h, 1 h and 2 h. The time 0 (T0) sample remained untreated. Cells were collected and subsequently lysed in RIPA buffer (Thermo Fisher Scientific) supplemented with cOmplete Protease Inhibitor and Benzonase. The lysate was cleared by centrifugation at 14,000g for 20 min at 4 °C. Protein concentration was determined by Bradford assay. For further immunoblot analysis, proteins were denatured in 2× SDS Sample Buffer by boiling at 95 °C for 5 min. Immunoblots were analysed and quantified using Fiji. Degradation half-time was determined by fitting the data in SigmaPlot 14.0 using an exponential decay function mentioned above.
Preparation of DNase I Sepharose beads
Cyanogen bromide-activated Sepharose 4B beads (1 g; Sigma-Aldrich) were washed up to 10 times with 1 mM HCl. 100 mg DNase I (Roche) was dissolved in 0.1 M NaHCO3 and 0.5 mM CaCl2. DNase I solution was added to washed Cyanogen bromide-activated Sepharose 4B beads and incubated at 4 °C with vertical rotation overnight. Unreacted groups in the resin were blocked upon incubation with 0.2 M ethanolamine pH 8.0 and 0.5 mM CaCl2 for 2 h at 4 °C. DNase I conjugated Sepharose 4B beads were washed with two alternating cycles of 0.1 M sodium acetate pH 4.5 and 0.1 M ethanolamine pH 8.0. Beads were stored in 10 mM Tris-HCl pH 7.4, 1 mM CaCl2, 10% glycerol, 1 mM DTT, 0.02% NaN3, cOmplete Protease Inhibitor at 4 °C.
DNase I pulldown
U-2 OS cells were lysed in lysis buffer (10 mM Tris-HCl pH 7.4, 1 mM CaCl2, 1 mM DTT, 0.2 mM ATP, 0.5% Triton X-100, 1 mM PMSF, cOmplete Protease Inhibitor, Benzonase) on an end-over-end rotor for 30 min at 4 °C. Lysate was cleared by centrifugation at 16,000g for 20 min at 4 °C. Protein concentration was determined by Bradford assay. DNase I conjugated Sepharose 4B beads were equilibrated by washing 3 times with DNase I binding buffer (10 mM Tris-HCl pH 7.4, 1 mM CaCl2, 10% glycerol, 1 mM DTT, 0.2 mM ATP, cOmplete Protease Inhibitor). Beads were centrifuged at 500g for 2 min at 4 °C and supernatant was discarded. Equilibrated DNase I conjugated Sepharose 4B beads were added to cleared lysate and incubated for 3 h at 4 °C with vertical rotation. Beads were washed once with DNase I washing buffer (10 mM Tris-HCl pH 7.4, 1 mM CaCl2, 10% glycerol, 1 mM DTT, cOmplete Protease Inhibitor), once with DNase I washing buffer supplemented with 0.3 M NaCl, and twice with DNase I washing buffer. For immunoblotting the bound protein was eluted in 2× SDS Sample Buffer by boiling at 95 °C for 5 min.
Ribosome isolation
U-2 OS cells were lysed in standard lysis buffer on an end-over-end rotor for 30 min at 4 °C. Lysate was cleared by centrifugation at 15,000g for 15 min at 4 °C to remove large debris. Pre-cleared lysate was further centrifuged at 330,000g for 1 h at 4 °C in a SW 55 Ti rotor (Beckman Coulter) to pellet ribosomes. The supernatant and pellet fractions were used for immunoblot analysis.
Polysome gradient analysis
Cells were treated with 100 µg ml−1 CHX and subsequently lysed in lysis buffer (5 mM Tris-HCl pH 7.4, 1.5 mM NaCl, 2.5 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, 0.5% sodium deoxylate, 100 µg ml−1 CHX, 100 U ml−1 SUPERase*In, cOmplete Protease Inhibitor, Benzonase). The RNA concentration of the pre-cleared lysate was determined by measuring absorbance at 260 nm, and a total RNA amount of 300 µg was loaded onto the gradients. Sucrose density gradients (10%–45%) were prepared in SW41 ultracentrifuge tubes (Steton) using a BioComp Gradient Master (BioComp Instruments) according to the manufacturer’s instructions. The individual 10% and 45% sucrose solutions were prepared in polysome buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 15 mM MgCl2, 1 mM DTT, 100 µg ml−1 CHX, cOmplete Protease Inhibitor). The gradients were centrifuged for 2.5 h at 40,000 rpm in a SW 41 Ti rotor (Beckman Coulter) at 4 °C. Polysome profiles were obtained using the Triax Flow Cell Firmware v.2.30.4.211. The gradients were fractionated using a piston gradient fractionator coupled to an A254 nm spectrophotometer (Biocomp). Polysome fractions were precipitated with 10% trichloroacetate and protein precipitates were washed with ice cold acetone. For immunoblotting the proteins were denatured in 2x SDS Sample Buffer by boiling at 95 °C for 5 min.
Solubility assay
U-2 OS cells were electroporated with a plasmid encoding either actin–SunTag–Xbp1u+ or actin(G150P)–SunTag–Xbp1u+. Twenty-four hours after electroporation cells were washed twice with ice cold PBS and lysed in RIPA buffer supplemented with cOmplete Protease Inhibitor and Benzonase. Protein concentration was determined by Bradford assay. The sample was divided into a total fraction and a fraction subjected to centrifugation at 20,000g for 20 min at 4 °C (pellet). The supernatant was transferred to a fresh tube, while the pellet was washed with RIPA buffer supplemented with cOmplete Protease Inhibitor and Benzonase, then centrifuged again under the same conditions. The total and supernatant fractions were denatured in 2× SDS Sample Buffer, while the pellet fraction was denatured in 1× SDS Sample Buffer by boiling at 95 °C for 5 min. Fractions were analysed by immunoblotting.
Single-molecule tracking in live cells
Fluorescence microscopy
Two-colour simultaneous imaging was performed on a Zeiss Elyra PS.1 inverted super-resolution microscope equipped with two Andor iXonEM+ DU-897D EM-CCD cameras and a custom-built image splitter with motorized xyz–rotation adjustment. Camera alignment was automatically performed with a built-in alignment pattern. All images were acquired using ZEN black 2.1 SP3 software using Zeiss alpha Plan-Apochromat 100×/1,46 Oil DIC oil immersion objective in TIRF (total internal reflection fluorescence) mode. The TIRF angle was set between 63° and 65°. The two fluorescence channels were acquired in parallel with the two cameras. Each time series consisted of 500 frames with 50 ms exposure time. During imaging, cells were incubated in a Tokai Hit stage-top incubator at 37 °C and 5% CO2.
Sparse labelling
Cells used for imaging were cultured in a glass-bottom dish (Ibidi, µ-Dish 35 mm, high glass bottom). Before labelling, cells were washed with Hanks’ Balanced Salt Solution (HBSS, Gibco, 14025092) and incubated with fresh culture media. Janelia Fluor (JF) HaloTag ligands (Promega) and SNAP-tag ligand TF-TMR dye (provided by K. Johnsson)34 were dissolved in DMSO to prepare stock solutions. Fluorescent ligands were added to the cells and incubated for 20–30 min. Excess dye was washed out by rinsing the cells three times with HBSS. Cells were incubated with fresh media for at least 5 min and washed again before imaging.
SPT and colocalization analysis
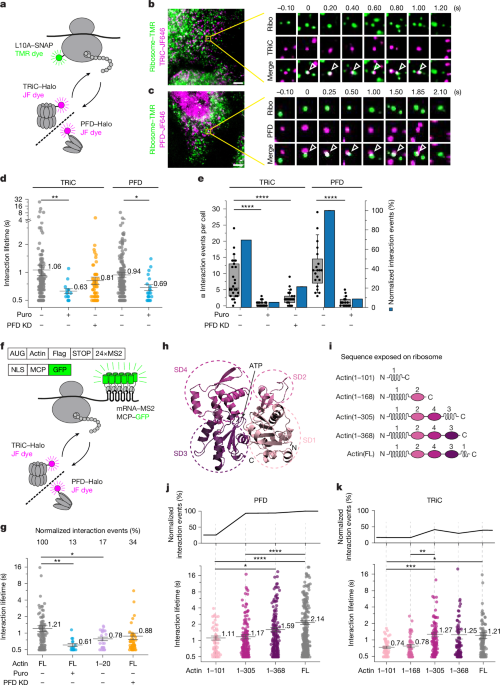
Tracking of individual particles was performed in Fiji59 with the plugin TrackMate60. Particles in each frame were detected using the Laplacian of Gaussian detector. Multiple trajectories were then determined from the detected particles using the simple linear assignment problem (LAP) tracker. The linking max distance, gap-closing max distance, and gap-closing max frame gap parameters were set to 0.5 µm, 0.5 µm and 0 frames, respectively. Trajectories detected in the two channels were analysed to identify colocalization events using KNIME as described61. Pairwise colocalization analysis was performed by measuring the distances from each molecule in the first channel to all the molecules in the second channel. Molecule pairs were classified as co-localization if their distance was less than 500 nm. Tracks with co-localized molecules that persisted for more than 500 ms were used to analyse the interaction lifetime between chaperones and substrates. A 500 ms threshold was applied to exclude short-lived contacts, which are dominated by non-specific encounters and obscure the detection of condition-specific differences, thereby enriching for biologically meaningful interactions. Mean square displacement was used to determine the diffusion coefficient of each track.
Statistical analysis of lifetime and interaction events
Welch’s ANOVA test was used for statistical comparison of measured lifetimes across more than two groups. This test is suitable for datasets that are not normally distributed and have unequal sample sizes, provided that each group contains more than 15 observations62,63. For comparison of two groups, the non-parametric Mann–Whitney U-test was applied. When comparing interaction events across more than two groups, a one-way ANOVA followed by Tukey’s post hoc test was used if a significant overall difference was detected (hereafter referred to as ANOVA).
Kinetic modelling of chaperone–substrate interactions
The interaction lifetime between chaperone and substrates was calculated from the duration of their co-movement events. To resolve distinct kinetic components, we fitted the 1 − CDF of the interaction lifetimes using a constrained nonlinear least-squares fit. Two kinetic models were evaluated: a single-component model (\(F(t)={{\rm{e}}}^{-t/\tau }\)) and a two-component model (\(F(t)=\alpha {{\rm{e}}}^{-t/{\tau }_{1}}+(1-\alpha ){{\rm{e}}}^{-t/{\tau }_{2}}\)). In the two-component model, τ1 and τ2 represent the lifetime of each component, and α and 1 − α are their respective fractions. The two-component model was only accepted if it showed both higher coefficient of determination (R2) than the single-component model and a statistically significant second population (α > 5%).
Fraction of fluorescently labelled molecules
To estimate the percentage of labelled Halo-tagged protein at certain concentrations of fluorescent ligand, cells were incubated with Halo ligand JF646 dye (Promega) at the following concentrations: 100 nM, 150 nM, 250 nM and 500 nM for cells expressing TRiC–Halo and 2 nM, 10 nM, 100 nM and 250 nM for cells expressing PFD–Halo. Images were acquired using a fluorescence microscope. Average fluorescence intensity was quantified at different dye concentrations. The highest dye concentrations corresponding to the plateau of normalized fluorescence intensity (250 nM for PFD–Halo and 500 nM for TRiC–Halo), as well as concentrations 2-fold higher (500 nM for PFD–Halo and 1 µM for TRiC–Halo), were used for in-gel fluorescence analysis. These samples were compared to those labelled with the dye concentrations used for SPT (800 pM for PFD–Halo and 50 pM for TRiC–Halo). Cells treated in same way were lysed in RIPA buffer supplemented with cOmplete Protease Inhibitor and Benzonase, and proteins were separated by SDS–PAGE as previously described. To minimize quantification bias, fivefold less total protein was loaded for lysates from samples labelled at saturated dye concentrations compared to those used for SPT. Fluorescence signals were detected using an Amersham Typhoon Biomolecular Imager and quantified with Fiji. Coomassie staining was performed as a loading control (Extended Data Fig. 1k).
Mass spectrometry
Sample preparation for total proteome analysis
Cell pellets were resuspended in 400 µl of SDC buffer containing 1% sodium deoxycholate (SDC; Sigma-Aldrich), 40 mM 2-chloroacetamide (Sigma-Aldrich), 10 mM tris(2-carboxyethyl)phosphine (TCEP, Thermo Fisher Scientific), and 100 mM Tris, pH 8.0. After incubation for 5 min at 95 °C, the samples were ultrasonicated for 10 min using 10 cycles of 30 s at high intensity with a 30 s pause between cycles (Bioruptor, Diagenode). The incubation and ultrasonication steps were repeated once more. The samples were then diluted 1:1 with MS-grade water (VWR), and proteins were digested with 1 µg Lys-C (Wako) for 4 h at 37 °C, followed by an overnight digestion at 37 °C with 2 µg trypsin (Promega). The resulting peptide solution was acidified with TFA to a final concentration of 1%, and then desalted using SCX-stage tips.
Liquid chromatography–mass spectrometry data acquisition
Liquid chromatography–mass spectrometry analysis was performed using an Easy-nLC 1200 (Thermo Fisher Scientific) nanoflow system coupled with a QExactive HF mass spectrometer (Thermo Fisher Scientific). Chromatographic separation was achieved on a 30-cm column (inner diameter: 75 µm; packed in-house with ReproSil-Pur C18-AQ 1.9 µm beads, Dr. Maisch GmbH). Peptides were injected onto the column in buffer A (0.1% (v/v) formic acid), with the column heated to 60 °C. Peptides were eluted at a flow rate of 250 nl min−1 using a gradient from 2% to 30% buffer B (80% acetonitrile, 0.1% formic acid) over 120 min (QExactive HF), followed by a ramp to 60% over 10 min, then to 95% over the next 5 min, which was maintained for another 5 min to measure the total proteome. For immunoprecipitation measurements, peptides were eluted using a gradient from 7% to 30% buffer B over 60 min, followed by an increase to 60% over 15 min, then to 95% over the next 5 min, and maintained at 95% for an additional 5 min. The QExactive HF mass spectrometer was operated in data-dependent mode, with survey scans acquired over the m/z range of 300–1,650 at a resolution of 60,000 at m/z = 200. Up to 10 of the top precursors were selected and fragmented using higher-energy collisional dissociation (HCD) with a normalized collision energy of 30. The AGC target for MS and MS2 scans were set to 3E6 and 1E5, respectively, with maximum injection times of 100 ms for MS and 60 ms for MS2. Dynamic exclusion was set to 30 s.
Mass spectrometry data analysis
Raw data were processed using the MaxQuant computational platform (v.2.2.0.0) with default settings. The peak list was searched against the human proteome database (UniProt: SwissProt and TrEMBL) as well as protein sequences of interest, with an allowed precursor mass deviation of 4.5 ppm and an allowed fragment mass deviation of 20 ppm. The default setting for individual peptide mass tolerances in MaxQuant was used in the search. Cysteine carbamidomethylation was set as a static modification, while methionine oxidation, N-terminal acetylation, deamidation on asparagine and glutamine, and phosphorylation on serine, threonine, and tyrosine were set as variable modifications. Protein quantification across samples was performed using the label-free quantification (LFQ) algorithm in MaxQuant, and iBAQ (intensity-based absolute quantification) values were calculated for each protein.
Selective ribosome profiling
Preparation of ribosome fractions for selective ribosome profiling
Cells were treated with 100 µg ml−1 CHX for 1 min at 37 °C before collection. Cells were washed two times with PBS containing Ca2+/Mg2+ and incubated with 2 mM DSP (dithiobis(succinimidyl propionate)) for 30 min at room temperature for crosslinking. DSP solution was removed and incubated with quenching buffer (20 mM Tris-HCl pH 7.5, 300 mM glycine) for 15 min at room temperature. Cells were washed 2 times with ice cold PBS containing Ca2+/Mg2+ and lysed in lysis buffer (20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 1% IGEPAL CA-630, 20 U ml−1 apyrase, 1 mM PMSF, 2 mM DTT, 100 µg ml−1 CHX, cOmplete Protease Inhibitor, benzonase) on an end-over-end rotor for 30 min at 4 °C. Lysate was digested with 500 U per 1 mg RNase I (Lucigen) at 4 °C for 1 h. Ribosome pellets were isolated using a 25% sucrose cushion prepared in lysis buffer without IGEPAL CA-630 and supplemented with 20 U ml−1 SUPERase*In (Invitrogen). Samples were centrifuged at 55,000 rpm in a SW 55 Ti rotor (Beckman Coulter) for 1 h at 4 °C. Pellets were resuspended in lysis buffer and used for anti-HaloTag pull down. Samples of the total RNA from the ribosome pellets were resuspended in TRIzol reagent (Invitrogen). HaloTag pull down was performed as described above. RPFs were recovered and libraries were prepared as described36. Libraries were excised from an 8% TBE polyacrylamide gel and sequenced on an Illumina NextSeq 500 or NovaSeq 6000 system.
Ribosome profiling data analysis
Data were analysed as previously described64. In brief, reads were trimmed and demultiplexed using an awk script. The UMI were extracted using umi-tools with the flag ‘–extract-method=regex –bc-pattern = “ ˆ(?P$”’, which serves to remove duplicated reads arising from PCR amplification. The remaining reads were mapped against a non-coding RNA library, including rRNA using Bowtie2 (v.2.4.2) with the parameters ‘-N 1 -L 15’. The remaining unaligned reads were mapped against the human genome (hg38) using STAR (v.2.7.10a) with parameters ‘–outFilterMismatchNmax 2 –quantMode TranscriptomeSAM GeneCounts –outSAMattributes MD NH –outFilterMultimapNmax 1’. P-site offsets were calculated using the R-package riboWaltz. Co-translational interactions of TRiC and PFD were identified as described previously17. In summary, genes were filtered with a coverage below an average of 0.5 reads per codon. Then, positional enrichments were calculated using a two-tailed Fisher’s exact test to compare TRiC–PFD enriched ribosome–nascent chain complexes to input fractions at each position along the coding sequence. This process yielded an enrichment score, defined as an odds ratio, which compared the expected ratio at a given position to the actual observed ratio. A Benjamini–Hochberg correction was applied to test for significance at each position. Interactions were considered valid when an enrichment was observed for at least five codons and the adjusted P value was below 0.05. For plotting binding profiles of chaperones with individual transcripts, one read was added at each position prior to the calculation of the odds ratio in order to avoid division by zero.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.