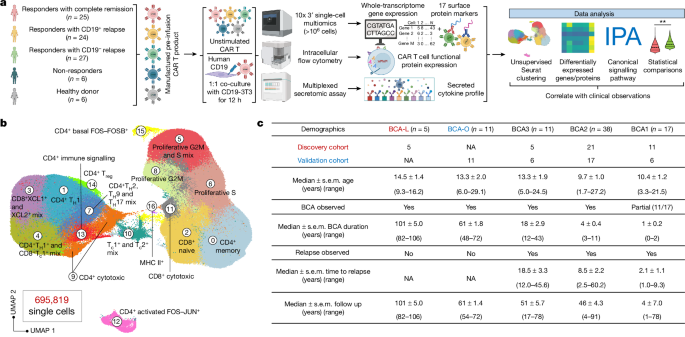
Samples from paediatric patients with ALL and HDs
The current study is a secondary investigation using patient samples collected from an existing clinical trial for which the University of Pennsylvania Institutional Board provided insight. Pre-infusion samples of CAR T cells were obtained from patients with relapsed/refractory B-ALL, participating in either a phase I/IIA pilot clinical trial designed to assess the safety and feasibility of CTL019 T cell therapy (ClinicalTrials.gov: NCT01626495) or a pilot study focusing on optimizing the timing of tocilizumab administration for managing CART19 therapy associated cytokine release syndrome (ClinicalTrials.gov: NCT02906371). Both trials were conducted jointly at the Childrenâs Hospital of Philadelphia and the University of Pennsylvania. Before participation, the patients or their guardians provided written informed consent in accordance with the principles outlined in the Declaration of Helsinki. Laboratory procedures adhered strictly to the guidelines established by the International Conference on Harmonization for Good Clinical Practice, employing standardized operating procedures and protocols for the receipt, processing, freezing and analysis of samples. Stringent ethical regulations were rigorously adhered to throughout the study. Primary T lymphocytes from HDs were provided by the University of Pennsylvania Human Immunology Core. To ensure compliance with HIPAA regulations, all of the samples were deidentified before analysis.
Generation of CTL019 cells
Autologous peripheral blood mononuclear cells were collected by standard leukapheresis. T cells were subsequently enriched through mononuclear cell elutriation, followed by thorough washing and activation using anti-CD3/CD28-coated paramagnetic beads. A lentiviral vector carrying a previously described CD19-specific CAR with a 4-1BB/CD3ζ transgene was constructed36, after which it was used to transduce the cells during the activation phase and was washed out 3âdays after the initiation of culture37. Cell expansion was facilitated using a rocking platform (WAVE Bioreactor System) for a duration of 8 to 12 days, and the beads were then magnetically removed. Finally, CTL019 cells were collected and cryopreserved for future use.
Generation of human-CD19-expressing NIH/3T3 cell line
We developed artificial APCs using mouse fibroblast NIH/3T3 cells to activate CAR T cells targeting the corresponding antigen. The NIH/3T3 cell lines were originally procured from the American Type Culture Collection (ATCC) and maintained in DMEM medium (Gibco, 11995-065) supplemented with 10% fetal bovine serum (FBS; Gibco, 16000044), within a humidified incubator set at 37â°C with 5% CO2. After reaching approximately 80% confluency, the cells were dissociated from the culture flask using 0.25% trypsin-EDTA (Gibco, 25200056) and transduced with a lentiviral vector encoding human CD19. Then, 3âdays after transduction, 1âÃâ106 cells were labelled with an antibody specific to the transduced epitope and sorted using the FACSAria (BD) sorter to achieve a purity exceeding 99% after the introduction of the transgene. Routine screening for mycoplasma contamination and authentication was conducted both before and after genetic modification. Subsequently, stable expressing clones were selected for expansion in T25 or T75 flasks and stored cryogenically for future use.
In vitro co-culture assay
CTL019 cells were thawed and cultured in OpTmizer T-Cell Expansion Basal Medium (Thermo Fisher Scientific, A1048501) supplemented with GlutaMAX Supplement (Thermo Fisher Scientific, 35050061) and 5% human serum AB (Gemini Bioproducts, GEM100-512) in a humidified incubator for an initial overnight rest on day 1. On day 2, dead cells were eliminated using the Dead Cell Removal Kit (Miltenyi Biotec, 130-090-101) according to the manufacturerâs instructions, and a specific number of cells was enumerated using a haemocytometer before initiating co-culture assays. For stimulation with CD19-3T3 cells (APC cells), 1âÃâ106 CTL019 cells were combined with an equivalent number of APC cells in 2âml of medium; for the assessment of unstimulated conditions, 1âÃâ106 cells were prepared. All suspensions were cultured in RPMI medium (Gibco, 11875-119) supplemented with 10% FBS in a tissue-culture-treated 24-well plate (Thermo Fisher Scientific) for 12âh in the incubator. For functional experiments evaluating the response of the CAR T cell population to type 2 cytokines, recombinant human IL-4, IL-5 or IL-13 (R&D Systems, 204-IL-010, 205-IL-010, 213-ILB-010, respectively) was added to the medium at the specified concentrations. After co-culture, the cells were collected by vigorous pipetting, and the cell suspension was passed through a 20âμm filter to remove clumps before staining with PE-labelled monoclonal anti-FMC63 single-chain variable fragment (scFv) antibody (CAR19) (Y45, ACRO Biosystems, FM3-HPY53) for 1âh at 4â°C. Subsequently, the cells were labelled with anti-PE MicroBeads UltraPure (Miltenyi Biotec, 130-105-639) and loaded onto a MACS column positioned within the magnetic field of a MACS Separator. CAR+ cells, magnetically retained within the column, were isolated as the positively selected fraction, while untransduced CARâ cells flowed through.
Sample hashing and staining with DNA-barcoded antibodies for CITE-seq
We used hashtag reagents for sample barcoding, enabling the amalgamation of eight samples into a single lane for subsequent demultiplexing during analysis. Specifically, for human samples, the hashtags consisted of two antibodies recognizing ubiquitous surface markers, CD298 and β2 microglobulin, each conjugated to the same oligonucleotide containing the barcode sequence. The magnetically selected CAR+ cells obtained from the preceding step, originating from four individuals, along with the corresponding basal unstimulated CAR T cells from each individual, underwent blocking using 5âµl of human TruStain FcX Fc Blocking reagent (BioLegend, 422302). Subsequently, they were incubated with 1âµl (0.5âµg) of the respective TotalSeq-B anti-human Hashtag antibodies 1â9 (BioLegend) for 30âmin at 4â°C. After staining, the samples were washed twice with 500âµl of cell staining buffer (BioLegend, 420201) and pooled into a single tube. The combined cells were then incubated in an antibody cocktail comprising 2âµl (1âµg) of each TotalSeq-B anti-human antibody (BioLegend) according to the manufacturerâs protocol. The panel comprised a selection of antibodies targeting various cell surface markers including CD4 (RPA-T4, 300565), CD8 (SK1, 344757), CD45RA (HI100, 304161), CD45RO (UCHL1, 304257), CD62L (DREG-56, 304849), FAS (DX2, 305653), CD127 (A019D5, 351354), CD28 (G043H7, 302961), CD27 (O323, 302851), CCR7 (G043H7, 353249), HLA-DR (L243, 307661), CD69 (FN50, 310949), PD-1 (EH12.2H7, 329961), TIM3 (F38-2E2, 345053), LAG-3 (11C3C65, 369337), CTLA-4 (BNI3, 369629) and TIGIT (A15153G, 372727).
scRNA-seq library preparation and sequencing
The scRNA-seq libraries were prepared using the Chromium Single-Cell 3â² Library and Gel Bead Kit v3.1 (10x Genomics, PN-1000268). Initially, 20,000 TotalSeq antibody-stained CAR T cells were suspended in PBS (Gibco, 14190-144) with 0.04% bovine serum albumin (BSA; Sigma-Aldrich, A7030) buffer and loaded onto the Chromium Next GEM chip G, where cells and uniquely barcoded beads were partitioned into nanolitre-scale gel beads-in-emulsion (GEMs). Within each GEM, cell lysis occurred, followed by reverse transcription of the released mRNA and isolation and amplification of the barcoded complementary DNA by PCR for 12 cycles. Subsequent separation of hashtag/surface protein oligo-derived cDNAs (<200âbp) and mRNA-derived cDNAs (>300âbp) was accomplished using 0.6Ã SPRI bead (Beckman Coulter) purification on cDNA reactions. After fragmentation, end repair and poly(A) tailing, sample indexes were incorporated, and amplification was performed. The final libraries underwent quality-control checks before being sequenced on the Illumina NovaSeq 6000 sequencing system, with paired-end reads of 150âbp in length. Three samples were pooled and sequenced per 800G flow cell at a gene and hashtag/surface protein library pooling ratio of 8:1.
scATAC and gene co-profiling library preparation and sequencing
Single-cell co-profiling of epigenomic landscape and gene expression in the same single nuclei was performed using the Chromium Next GEM Single Cell Multiome ATAC + Gene Expression kit (10x Genomics, PN-1000283). Initially, CAR+ cells stimulated with CD19-3T3 from 6 patients, with or without a 10ângâmlâ1 IL-4 supplement, underwent washing, counting and nucleus isolation, with an optimized lysis time of 3âmin. Subsequently, the isolated nucleus suspensions were incubated in a transposition mix containing a transposase enzyme, facilitating preferential fragmentation of DNA in open chromatin regions. Concurrently, adapter sequences were introduced to the ends of the DNA fragments. Approximately 9,250 nuclei were then loaded onto the Chromium Next GEM Chip J to target a final recovery of around 6,000 nuclei. During GEM generation, gel beads introduced a poly(dT) sequence that enables production of barcoded, full-length cDNA from mRNA for gene expression profiling and a spacer sequence facilitating barcode attachment to transposed DNA fragments for ATAC profiling. After GEM incubation, purification and pre-amplification PCR, separate ATAC and gene libraries were constructed using the standard protocol. After quality assessment, both libraries underwent paired-end 150âbp read sequencing on the Illumina NovaSeq 6000 sequencing system, achieving an average depth of 24,305 read pairs per nucleus for the ATAC library and 13,756 read pairs per nucleus for the gene library.
Intracellular cytokine detection assay for CD19-3T3-stimulated patient CAR T cells
The co-cultured cells underwent a series of processing steps for immunostaining. Initially, they were washed twice in PBS and then stained for 20âmin at room temperature with Live Dead Blue detection reagent (Thermo Fisher Scientific, L34962), diluted to 1:800 in PBS, to assess cell viability. Next, cells were washed twice in FACS staining buffer and subsequently stained for surface molecules for 20âmin at room temperature. To fix the stained cells, the Cytofix/CytoPerm Fixation/Permeabilization Kit (BD Biosciences, 554714) was used for 20âmin at room temperature, while being protected from light. Subsequently, cells were washed twice with 1Ã perm/wash buffer and then stained for CAR19 and intracellular cytokines using antibodies in perm/wash buffer. This staining process was performed for 20âmin at room temperature in the dark. Cells then underwent two additional washes with perm/wash buffer before being resuspended in FACS staining buffer for subsequent analysis. The following antigens were stained using the specified antibody clones: anti-FMC63 scFv (Y45, ACRO Biosystems, FM3-HPY53), CD3 (SK7, BD Biosciences, 564001), CD4 (OKT4, BioLegend, 317442), CD8a (RPA-T8, BioLegend, 301042), CD19 (HIB19, BD Biosciences, 561121), CD14 (M5E2, BD Biosciences, 561391), IL-3 (BVD3-1F9, BioLegend, 500606), IL-4 (MP4-25D2, BioLegend, 500834), IL-5 (TRFK5, BioLegend, 504306), IL-13 (JES10-5A2, BioLegend, 501916) and IL-31 (1D10B31, BioLegend, 659608). Cell-surface antibodies were used at a 1:100 dilution during staining, and intracellular antibodies at a 1:50 dilution. The samples were analysed on the Cytek Aurora flow cytometer, and data analysis was conducted using FlowJo v.10.8.0.
Multiplexed secretomic assay
After stimulation with CD19-3T3 cells, around 30,000 magnetically enriched CAR+ cells were processed for membrane staining (IsoPlexis, STAIN-1001-1). Subsequently, these cells were loaded onto the IsoCode chip (IsoPlexis, ISOCODE-1001-4), comprising 12,000 chambers prepatterned with an array of 32 cytokine capture antibodies. The chip was further incubated in the IsoLight machine for 16âh at 37â°C with 5% CO2 supplementation. A cocktail of detection antibodies was then applied to detect the secreted cytokines, followed by fluorescence labelling. The resulting fluorescence signals were analysed using IsoSpeak v.2.8.1.0 (IsoPlexis) to determine the numbers of specific cytokine-secreting cells and the intensity level of each cytokine. For downstream analyses, the raw data pertaining to type-2-related cytokines, including IL-4, IL-5, IL-9, IL-10, IL-13 and IL-21, were extracted.
Generation of STAT6 and GATA3 knockdown CAR T cells
To generate STAT6 and GATA3 knockdown CAR T cells, lentiviral particles containing short hairpin RNA against STAT6 (shSTAT6) and shGATA3 were first produced in HEK293T cells. The cell lines were originally obtained from ATCC and tested negative for mycoplasma contamination. These cells were transfected with plasmids encoding pCMV-VSV-G (Addgene, 8454), pCMV-dR8.2 dvpr (Addgene, 8455), and either pLKO.1-puro_shSTAT6 (pLKO.1-puro, Addgene, 8453) or pLKO.1-puro_shGATA3, using the calcium phosphate transfection method. CAR T cells were subsequently spin-transduced with shSTAT6 or shGATA3 lentivirus particles on two consecutive days to ensure efficient transduction. The pool of shSTAT6 or shGATA3 sequences was as follows:
shSTAT6-1: 5â²-CCGGAGCGGCTCTATGTCGACTTTCCTCGAGGAAAGTCGACATAGAGCCGCTTTTTTG-3â²; shSTAT6-2: 5â²-CCGGAGCACCCTTGAGAGCATATATCTCGAGATATATGCTCTCAAGGGTGCTTTTTTG-3â²; shGATA3-1: 5â²-CCGGAGCCTAAACGCGATGGATATACTCGAGTATATCCATCGCGTTTAGGCTTTTTTG-3â²; shGATA3-2: 5â²-CCGGCCCAAGAACAGCTCGTTTAACCTCGAGGTTAAACGAGCTGTTCTTGGGTTTTTG-3â². The resulting CAR T cells were expanded for an additional 2âdays before use. The knockdown efficiency was assessed at both the gene and protein expression levels to confirm the efficacy of STAT6 and GATA3 silencing.
Serial proteomic profiling of patient serum samples
The Cytokine Human Magnetic 30-Plex Panel (Invitrogen, LHC6003M) was used to detect serum proteins in a cohort comprising 33 patients in our discovery cohort. Serum samples, cryopreserved at â80â°C, spanning from 2âdays before to 63 days after CTL019 infusion, were thawed and analysed according to the manufacturersâ protocols. Measurements were conducted using the FlexMAP 3D instrument (Luminex), and data acquisition and analysis were performed using xPONENT software (Luminex). Moreover, the Olink Explore 384 panel (Olink Proteomics) was used to measure serum proteins in eight patients from our validation cohort, with all protein data reported in normalized expression values on a log2 scale. In the discovery cohort, serum collections may have been performed on different days for various patients within a given time frame. In the validation cohort, serum collections were consistently conducted for all patients on specified days.
Mice and tumour cell lines
NOD/SCID/IL-2Rγnull (NSG) mice (aged 6âweeks) were procured from Charles River Laboratory. All mice were housed in the Center of PhenoGenomics (CPG) animal facility at EPFL, kept in individually ventilated cages at 19â23â°C with 45â65% humidity and maintained under a 12âhâ12âh darkâlight cycle. All experimental procedures involving mice were ethically approved by Swiss authorities (Canton of Vaud, animal protocol ID 3533) and adhered to the guidelines set forth by the CPG of EPFL. Nalm6 cell lines, sourced from ATCC, were screened and confirmed to be free of mycoplasma contamination. These cells were stably transduced with GFP-luciferase lentivirus for downstream experimentation. Culturing of Nalm6 cells was conducted in RPMI medium supplemented with 10% FBS and 200âUâmlâ1 penicillinâstreptomycin (Gibco, 15140122).
In vivo xenograft mouse studies
A total of 1âÃâ106 Nalm6-luciferase cells were i.v. injected into NSG mice to establish the leukaemia xenograft mouse model. Mice were randomized after tumour injection before initiating treatment. Then, 1âweek later, 2âÃâ106 CAR T cells were adoptively infused through tail vein injection. Tumour growth was monitored weekly using the Xenogen IVIS fluorescence/bioluminescence imaging system (PerkinElmer). In brief, mice were intraperitoneally injected with bioluminescent substrate d-luciferin potassium salt (150âmg per kg; Abcam, ab143655). Then, 10âmin after injection, the mice were anaesthetized and subjected to the luminescent imaging system to quantify tumour burden. The surviving mice were rechallenged with 1âÃâ106 Nalm6-luciferase cells 17âdays after the CAR T infusion. Mice were euthanized when body weight loss was beyond 15% of the baseline weight, or any signs of discomfort were detected by the investigators or as recommended by the veterinarian who monitored the mice every other day.
In vitro repeat stimulation assay
CTL019 cells were thawed and allowed to rest for 3â4âh before undergoing sequential staining with PE-labelled monoclonal anti-FMC63 scFv antibody and anti-PE MicroBeads, according to previously established protocols. CAR+ cells, magnetically enriched using the MACS column, were then co-cultured with Nalm6 cells in a six-well plate at specified E/T ratios. Throughout the assay period, the number of CAR T cells and Nalm6 cells was assessed daily using flow cytometry. Additional Nalm6 cells were added as necessary to maintain the designated E/T ratio. Each evaluated condition was prepared with 3 or 4 technical replicates.
Enhanced type 2 CAR T cell manufacture
The manufacturing process for enhanced type 2 CAR T cells largely adhered to previously established protocols, with a notable modification involving the addition of either 10ângâmlâ1 or 50ângâmlâ1 of IL-4 (R&D Systems, 204-IL-010) throughout the entire culture and expansion process. This supplementation was introduced in addition to the standard inclusion of 5ângâmlâ1 each of IL-7 (Miltenyi Biotec, 130-095-367) and IL-15 (Miltenyi Biotec, 130-095760) used in traditional manufacturing procedures.
Flow cytometry
Mouse blood (50âμl) was collected from the tail at specified timepoints for peripheral CAR T cell analysis. The collected samples were resuspended in PBS with EDTA (2âmM), and the red blood cells were removed using ACK lysis buffer (Gibco, A1049201). For surface marker staining, cells were incubated with an antibody panel at 4â°C for 30âmin, followed by live/dead staining. Cells were then washed and resuspended in PBS with 0.2% BSA for flow cytometry analysis. Intracellular cytokine staining was performed by first stimulating cells with a cell stimulation cocktail (Invitrogen, 00-4970-03) for 5âh at 37â°C to induce cytokine production. Subsequently, cells were stained for surface markers and live/dead dye as previously described, then fixed and permeabilized using the Cytofix/Cytoperm Kit (BD Biosciences). Intracellular staining with the indicated antibody panel was conducted according to the manufacturerâs protocol. Data were collected using the Attune NxT Flow Cytometer with Attune NxT Software v.3 (Invitrogen) and analysed using FlowJo v.10.6.1 (Tree Star). Gate margins were determined by isotype controls and fluorescence-minus-one controls.
The following antibodies, each with their specified clones, were procured from BioLegend and used for flow cytometry analysis in both in vitro and in vivo functional assays: FAS (DX2, 305624), IL-13 (JES10-5A2, 501916), CD27 (LG.3A10, 124249), CD45RO (UCHL1, 304238), TNF (MAb11, 502940), CD3 (OKT3, 317306), GZMB (GB11, 515403), CD223 (LAG-3) (11C3C65, 369312), CD366 (TIM3) (F38-2E2, 345016), CD197 (CCR7) (G043H7, 353235), IL-4 (MP4-25D2, 500832), CD19 (HIB19, 302216), CD4 (OKT4, 317416), IL-5 (TRFK5, 504306), CD8 (SK1, 344724), CD279 (PD-1) (EH12.2H7, 329952), KLRG1 (MAFA) (2F1/KLRG1, 138426), IFNγ (4S.B3, 502530) and the Zombie Aqua Fixable Viability Kit (423102). Monoclonal anti-FMC63 antibody (Y45, FM3-HPY53) was purchased from ACRO Biosystems. Cell-surface antibodies were used at a 1:100 dilution during staining, intracellular antibodies at a 1:50 dilution, and live/dead staining at a 1:1,000 dilution.
Single-cell transcriptome data processing and analysis
A total of 44 paired scRNA-seq and CITE-seq libraries were sequenced; detailed data quality metrics are provided in Supplementary Table 2. The sequencing data underwent alignment to the GRCh38 human reference genome, followed by barcode and unique molecular identifier counting, ultimately generating a digital gene expression matrix using Cell Ranger v.6.1.2 (10x Genomics). The subsequent data analysis was conducted according to the Seurat v.4 pipeline38. The hashtag oligos expression was used to demultiplex cells back to their original sample of origin, while also identifying and excluding cross-sample doublets. Cells flagged as doublets (two barcodes detected) or lacking barcodes were omitted from the analysis. Only cells expressing a gene count ranging from 200 to 7,000 and exhibiting less than 10% mitochondrial gene content were retained for downstream analysis. For the whole-dataset analysis (Fig. 1b), a random grouping approach was implemented in which every 4 libraries were combined and designated as a single batch, resulting in 11 different batches. Subsequently, a fast integration method named reciprocal principal component analysis (PCA)39 was used with the default parameters to mitigate potential batch effects and enable large-scale data integration. This involved splitting the dataset into Seurat objects based on sequencing batch, independent normalization and variable feature identification for each dataset. Integration and PCA were conducted on repeatedly variable features across datasets, with anchors identified using the FindIntegrationAnchors function and subsequent dataset integration with IntegrateData. Standard workflows for visualization and clustering were then implemented. For subclustering analyses of basal unstimulated or CD19-3T3-stimulated CAR T cells from both the discovery and validation cohorts, the sctransform normalization method in Seurat was used40. This method is specifically designed to capture sharper biological heterogeneities in scRNA-seq datasets, with no significant batch effects observed for these analyses. DEGs were identified using the FindMarkers function for pairwise comparisons between cell groups or clusters, applying a log-transformed fold change threshold of 0.25 to select significant genes. Moreover, module scores based on predefined gene sets were computed using the AddModuleScore function.
scATAC and gene co-profiling data processing and analysis
Cell Ranger ARC v.2.0.2 (10x Genomics) was used to perform sample demultiplexing, barcode processing, identification of open chromatin regions, and simultaneous counting of transcripts and peak accessibility in single cells from the sequenced data. The output per barcode matrices underwent joint RNA and ATAC analysis using Signac (v.1.12.0)41 and Seurat (v.4)38. Per-cell quality control metrics were computed, including the nucleosome banding pattern (stored as nucleosome_signal) and the transcriptional start site (TSS) enrichment score for the ATAC component. These metrics were used to identify and remove outliers, with the quality report of each sample meticulously documented in Supplementary Table 4. Quality filtering criteria adhered to the default settings. Specifically, cells were retained if they exhibited an ATAC peak count ranging from 1,000 to 100,000, a gene count ranging from 1,000 to 25,000, a nucleosome_signal below 2 and a TSS enrichment score exceeding 1. To enhance the accuracy of peak identification, we used MACS2 v.2.2.9.1 with the CallPeaks function, a widely used tool for peak calling in chromatin accessibility analysis42. Subsequently, we constructed a joint neighbour graph representing both gene expression and DNA accessibility measurements using weighted nearest-neighbour methods in Seurat v.4. To investigate potential regulatory elements for genes of interest, we used the LinkPeaks function. This method identifies sets of peaks that may regulate gene expression by computing the correlation between gene expression and accessibility at nearby peaks, while correcting for biases due to GC content, overall accessibility and peak size. In preparation for motif analyses, we used the AddMotifs function to integrate DNA sequence motif information into the dataset. Furthermore, we computed a per-cell motif activity score using chromVAR43. For footprinting analysis of motifs with positional information, we used the Footprint function to gather and store all necessary data within the assay.
LâR interaction analysis
The R toolkit Connectome (v.1.0.0)44 was used to investigate cellâcell connectivity patterns using ligand and receptor expression values from our scRNA-seq datasets with the default parameters. The normalized Seurat object served as input, and cluster identities were used to define nodes in the interaction networks, resulting in an edge list connecting pairs of nodes through specific LâR mechanisms. We selected top-ranked interaction pairs for visualization, prioritizing those that are more likely to be biologically and statistically significant based on the scaled weights of each pair. The thickness of edges is directly proportional to correlation weights, with wider edges indicating a higher level of interaction. The sources.include and targets.include parameters were used to specify the source cluster emitting ligand signals and the target cluster expressing receptor genes that sense the ligands.
Ingenuity pathway analysis
Ingenuity Pathway Analysis (Qiagen)45 was used to identify the underlying signalling pathways regulated by the DEGs characterizing each identified cluster or response group. To achieve this, the DEG list, along with corresponding fold change values, P values and adjusted P values for each gene, were loaded into the dataset. Using the Ingenuity knowledge base (genes only) as a reference set, core expression analysis was performed. T-cell-related signalling pathways were specifically selected from the identified canonical pathways to represent the primary functional profile of each group. The activation or inhibition level of specific pathways was determined using the z-score metric. Conceptually, the z-score serves as a statistical measure, assessing how closely the actual expression pattern of molecules in our DEG dataset aligns with the expected pattern based on literature for a particular annotation (zâ>â0, activated/upregulated; zâ<â0, inhibited/downregulated; zââ¥â2 or zââ¤ââ2 can be considered to be significant). The significance of each identified signalling pathway was determined using the right-tailed Fisherâs exact test, with the P value reflecting the probability of association between molecules from our scRNA-seq dataset and the canonical pathway reference dataset.
Statistical analysis
Statistical analyses were conducted using Prism v.10 (GraphPad) or R v.4.3.1. Unless otherwise specified, data are presented as meanâ±âs.e.m. For comparisons involving three or more groups, we used one-way ANOVA with Tukeyâs multiple-comparison test. For comparisons between two groups, two-tailed MannâWhitney U-tests were used for nonparametric data, two-tailed Studentâs t-tests for parametric data and two-tailed Wilcoxon matched-pairs signed-rank tests for paired nonparametric data. For single-cell-level comparisons, typically depicted in violin plots (as shown in Extended Data Fig. 7b,j), the normalized expression value of the top 10% single cells from each patient in each BCA group was included in the comparison.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.