Antibodies and reagents
The following antibodies were used in this study: RIPK1 (Cell Signaling Technology, catalogue no. 3493); p-hRIPK1 (Cell Signaling Technology, catalogue no. 65746); hRIPK3 (Cell Signaling Technology, catalogue no.13526); p-hRIPK3 (Cell Signaling Technology, catalogue no. 93654); hMLKL (Cell Signaling Technology, catalogue no. 14993; Abcam, ab183770; Thermo Fisher Scientific, PA5-34733); p-hMLKL (Cell Signaling Technology, catalogue no. 91689); anti-SIGLEC12 (Boster, A10550-1; Invitrogen, PA5-110369; Invitrogen, PA5-31457); TMPRSS4 (Cell Signaling Technology, catalogue no. 84382); anti-HMGB1 (Cell Signaling Technology, catalogue no. 6893); anti-β-actin (Sigma-Aldrich, A5316); anti-FLAG (Sigma-Aldrich, F1804); anti-FLAG–HRP (Cell Signaling Technology, catalogue no. 86861); anti-mouse IgG (Cell Signaling Technology, catalogue no. 7074); anti-rabbit IgG (Cell Signaling Technology, catalogue no. 7076), anti-mouse IgG (H + L) and F(ab′)2 Fragment (Alexa Fluor 488 Conjugate) (Cell Signaling Technology, catalogue no. 4408). All primary antibodies were used at 1:2,000 dilution. All secondary antibodies were used at 1:5,000 dilution.
The following reagents were used in this study: SM-164 (S7089), Nec-1s (S8641), GSK′872 (S8465), NSA (S8251), emricasan (S7775) and erastin (S7242) from SelleckChem; etoposide (E1383), α-ketoglutarate (349631), lipopolysaccharide (L4391), luminol (A8511), p-coumaric acid (C9008) and anti-FLAG M2 agarose beads (M8823) from Sigma-Aldrich; nigericin (11437) from the Cayman Chemical Company; Lipofectamine 3000 (L3000015), SuperSignal West Atto Ultimate Sensitivity Substrate (A38556) and Prolong Diamond Antifade Mountant with DAPI (P36966) from Thermo Fisher Scientific; polyethylenimine (PEI; 24765-100) from Kyfora Bio; polybrene (TR-1003) from EMD Millipore; and 10X Tris/Glycine/SDS Electrophoresis Buffer (1610772), Tween 20 (1610781), Stacking Gel Buffer for PAGE (1610799), Resolving Gel Buffer for PAGE (1610798), Precision Plus Protein Dual Color Standards (1610394) and nitrocellulose membrane (1620115) from Bio-Rad. Autoradiography films were from MTC Bio (A8815).
Cell lines and growth media
The 293T, HEK293-FlpIn-TREx, HeLa and MEF cells were grown in DMEM (Cytiva, SH30243.FS), with l-glutamine, 4.5 g l−1 glucose and pyruvate; HT-29 cells were grown in McCoy’s 5A medium (Gibco, 16600-082, with l-glutamine); and A549, Jurkat and THP-1 cells were grown in RPMI 1640 medium (Fisher Scientific, SH30255.01, with HEPES and l-glutamine). All media were supplemented with 10% fetal bovine serum (GeminiBio), 1× non-essential amino acids (Cytiva, SH30238.01) and 1× antibiotic/antimycotic solution (Sigma-Aldrich, A5955). All cell lines were regularly tested for mycoplasma contamination using Lonza’s MycoAlert Kit (catalogue no. LT07-318) and tested negative for mycoplasma.
Human tumour and normal tissue samples
Deidentified human benign (normal) lung and colon and lung and colon adenocarcinoma tumour specimens were obtained from the University of Texas Southwestern Tissue Management Shared Resource. Patients were enrolled and consented to a protocol approved by the institutional review board. Formalin-fixed tissues were processed as in the ‘Immunohistochemistry of tissue slices’ section.
Molecular cloning and plasmids
Molecular cloning was performed using New England Biolabs restriction enzymes and T4 DNA ligase. Plasmids were transformed into chemically competent NEB Stable (for lentiviral plasmids, at 30 °C) and DH5α Escherichia coli cells (for non-lentiviral plasmids at 37 °C). Plasmid purification and extraction were performed using a QIAprep Spin Miniprep Kit (Qiagen, 27106) and QIAquick Gel Extraction Kit (Qiagen, 28704). sgRNAs targeting SIGLEC12 were cloned into pSpCas9(BB)-2A-GFP (pX458) (Addgene, catalogue no. 48138). Cloned plasmids were amplified and purified using a ZymoPURE II Plasmid Midiprep Kit (Zymo Research, D4201).
Generation of knockout cell lines using CRISPR–Cas9
Cells were transfected with the pSpCas9(BB)-2A-GFP (pX458) plasmids for 24 h using Lipofectamine 3000. Cells expressing the highest levels of GFP were then sorted into 96-well plates containing 150 µl of high-glucose DMEM or McCoy’s medium supplemented with 16% fetal bovine serum, 1× non-essential amino acids and 1× antibiotic/antimycotic. All flow cytometry experiments were performed at the UT Southwestern Cell Sorting Facility. Fluorescence-activated cell sorting was performed with a BD FACSAria II flow cytometer under sterile conditions using a 100 nozzle at 37 °C. About 2–3 weeks later, the clones were expanded into 12-well plates. Cells were lysed in 2× SSB (150 mM Tris-HCl, pH 6.8, 4% SDS, 0.01% bromophenol blue, 20% glycerol) + 2% β-mercaptoethanol (β-ME), and lysates were analysed for knockout screening using immunoblotting.
Generation of knockdown cell lines using lentiviral short hairpin RNA
MISSION short hairpin RNA plasmids targeting SIGLEC12, TMPRSS4 and MLKL were from Sigma-Aldrich. For generation of lentiviral particles, plasmids were transfected into 293T cells using PEI (3 μl per 1 μg DNA). Pseudoviral particles were collected 72 h after transfection, and cells were transduced in the presence of polybrene (8 μg ml−1). Cells were selected with puromycin (2 μg ml−1) for 48 h after transduction, and knockdown of target proteins was confirmed by immunoblotting after all cells in the untransduced control plates had been selected out.
Transfection experiments
Cells were transfected with a total of 1 µg of plasmid DNA or 3 µg of plasmid DNA in a 24-well plate or 100 mm dish, respectively. The ratio of pcDNA5-SIGLEC12–FLAG to pLenti-III-EF1-TMPRSS4–HA plasmid DNA was 1:1, and the total amount was adjusted using an empty vector. HeLa-RIPK3–HA cells were transfected using Lipofectamine 3000 (1.5 μl per 1 μg plasmid DNA), whereas 293T cells were transfected using PEI (3 μl per 1 μg plasmid DNA).
Cell death and viability assays
Necroptosis was induced with TSE cocktail (TSE; T: 30 ng ml−1 hTNF, S: 0.2 µM SM-164, E: 5 µM emricasan; 1 h pretreatment for S + E) for 8 h. Extrinsic apoptosis was induced with T and S (1 h pretreatment with S). Intrinsic apoptosis was induced with 100 µM etoposide. Pyroptosis was induced with α-ketoglutarate (15 mM) or LPS (1 μg ml−1, 4 h pretreatment) + nigericin (20 μM). Ferroptosis was induced with erastin (20 μM). CellToxGreen (Promega, G8731), ToxiLight Non-destructive Cytotoxicity BioAssay (Lonza, LT07-117) and CytoTox 96 Non-Radioactive Cytotoxicity Assay (LDH assay, Promega, G1780) were used for detection of cell death. Cell viability was measured using a CellTiter-Glo Luminescent Cell Viability Assay (Promega, G7570). For CellToxGreen, dye (used at 1:3,000 dilution) was added to the wells immediately before the fluorescence reads. For the Toxilight assay, 12.5 µl of culture medium was collected in triplicate and mixed with an equal volume of Toxilight reagent in 384-well plates. LDH assay was performed according to the manufacturer’s instructions, and absorbance was measured at 490 nm. For the cell survival assay, CellTiter-Glo reagent was added directly to the medium. Cells were lysed for 10 min in the dark at 25 °C, and luminescence was measured. Cell survival was normalized to that of an untreated control.
Live-cell imaging of cell death using Incucyte S3
Cells were seeded into 24-well plates and treated the following day. Plates were imaged using an Incucyte S3 Live-Cell Analysis System (Sartorius) with scans every 1 h using a ×10 objective, capturing phase contrast under the AI Scan module. Quantification of cell death was performed using the integrated AI Cell Health Analysis module, which applies a deep learning model to distinguish living and dead cells on the basis of morphology. Cell death was expressed as the percentage of dead cells per total cell count of cumulative cell counts over time.
Sample preparation for immunoblotting
Cells were lysed in 200 µl of 2× SSB + β-ME buffer. The plates were heated at 90 °C for 3 min, then cooled to room temperature for 5 min, and 1–2 µl of 1× benzonase (1:10 diluted with 50% glycerol from 10× supplier stock to obtain 1×; Santa Cruz Biotechnology, sc-202391) was added to the lysates to degrade genomic DNA for 5 min at room temperature, on a rocker. Total protein levels were normalized using a BCA kit (Thermo Fisher Scientific, 23225) or reducing-agent-compatible BCA kit (Thermo Fisher Scientific, 23250). For non-reducing sample preparation, cells were lysed in NLB buffer (NP-40 lysis buffer: 25 mM HEPES (pH 7.5), 0.2% NP-40, 120 mM NaCl, 0.27 M sucrose, 5 mM EDTA, 5 mM EGTA, 50 mM NaF, 10 mM b-glycerophosphate, 5 mM sodium pyrophosphate, 1 mM Na3VO4 (fresh), 0.1% β-ME, 1 mM phenylmethylsulfonyl fluoride (fresh), 2× complete protease inhibitor cocktail (Roche, 80024400)). Cell lysates were mixed with 4× SDS sample buffer without β-ME (non-reducing). Equal amounts of protein were resolved by SDS–PAGE and analysed using the indicated antibodies.
Immunoblotting
Total cell lysates or pull-down samples were heated at 90 °C for 5 min or 10 min in 2× or 4× SSB buffer, respectively, then subjected to 10% or 15% SDS–PAGE. Proteins were electrotransferred on to nitrocellulose membranes for 1.5 h at 0.4 A with the wet transfer tank submerged in an ice bath. The membranes were blocked for 1 h in TBST buffer containing 5% (w/v) skimmed milk and then incubated with the primary antibodies in TBST containing 5% (w/v) bovine serum albumin + 0.05% NaN3 overnight at 4 °C. Detection was carried out using HRP-conjugated secondary antibodies and a homemade chemiluminescence reagent (2.5 mM luminol, 0.4 mM p-coumaric acid, 100 mM Tris-HCl, pH 8.6, 0.018% H2O2).
Immunoprecipitation
Cells were seeded into a 100-mm dish at 50% confluence. After 24 h, cells were transfected with total 3 µg of plasmid DNA using 9 μl of PEI or 4.5 µl of lipofectamine 3000. After 14–24 h, cells were lysed in NRP buffer. Cell lysates were incubated and precipitated with specific primary antibody for overnight at 4 °C and then incubated with protein A/G-magnetic beads (Thermo Fisher Scientific, 88802) or anti-FLAG agarose beads (Sigma-Aldrich, M8823) for 4 h at 4 °C. Bound proteins were removed by boiling in 2× SSB + β-ME buffer for 10 min at 90 °C, separated by SDS–PAGE and immunoblotting, and visualized using a homemade chemiluminescence reagent.
Time-lapse live-cell imaging microscopy
Cells were seeded on a 24-well plate (Cellvis, P24-1.5 P). After 48 h, cells were treated with TSE with added CellToxGreen. Live-cell imaging was performed using a Nikon Spinning Disk Confocal CSU-W1 with a ×40 air objective. Both bright-field and GFP fluorescence channels were captured every 30 min for 24 h. During the live-cell imaging process, cells were maintained at 37 °C and 5% CO2.
Immunofluorescence microscopy
Cells were seeded on 70% ethanol-sterilized glass coverslips (AmScope, CS-R18-100). After 24 h, cells were transfected with total 0.5 µg of plasmid DNA using 0.75 µl of lipofectamine 3000. Cells were treated with TSE and then fixed in 4% paraformaldehyde for 12 min. Cells were washed twice with phosphate-buffered saline (PBS), with or without permeabilization using 0.05% Triton X-100 for 5 min. After incubation in a blocking buffer (1% bovine serum albumin in PBS) for 1 h, the cells were incubated overnight at 4 °C with the following primary antibodies: anti-FLAG or anti-HA. They were then incubated with the following Alexa Fluor secondary antibodies for 1 h at room temperature. Cells were mounted using Prolong Diamond Antifade Mountant with DAPI. Images were obtained with Laser scanning confocal Zeiss LSM880 inv. + Airyscan confocal, magnification 63x. Images were analysed using Fiji/ImageJ. Images are representative of at least ten fields of view per each sample.
Immunohistochemistry of tissue slices
Immunohistochemical analyses were conducted using a Dako Autostainer Link 48 system. Initially, the slides were baked at 60 °C for 20 min, followed by deparaffinization and hydration. Heat-induced antigen retrieval was performed using the Dako PT Link. The tissue samples were treated with a peroxidase block, and antibody incubations were carried out at a 1:200 dilution. Staining was visualized with a Nikon Widefield Epi-scope, magnification ×60. For Extended Data Fig. 4c, images were quantified using Fiji. In brief, the total tissue area was manually outlined, not considering large blood vessels. Then, the raw data was colour-deconvolved using the Colour Deconvolution2 plugin and thresholded manually to include the darkest SIGLEC12 staining. Results were expressed as the percentage area of SIGLEC12 relative to total tissue area.
Cell plug preparation
Cells were seeded in 100-mm culture dishes at approximately 80% confluence. After 24 h, cells were rinsed with 10 ml of cold PBS and fixed in 10 ml of 4% paraformaldehyde for 20 min at room temperature in the dark. Cells were then washed again with cold PBS and gently scraped into 1 ml of PBS. The suspension was centrifuged at 300g for 5 min at room temperature, and the pellet was resuspended in 100 μl of PBS and kept on ice. Separately, 20 ml of 1% (w/v) agarose in PBS was prepared by boiling and then cooled. A 500-μl aliquot of molten agarose was transferred to a sterile 2-ml microcentrifuge tube, and 100 μl of the cell suspension was added. The mixture was gently inverted 20 times to ensure even distribution, then centrifuged at 300g for 5 min at room temperature. The resulting cell plugs were maintained on ice for 1 h to solidify and stored at 4 °C for up to 2 days before processing for sectioning, haematoxylin and eosin staining, and preparation of unstained slides.
Electron microscopy
For transmission electron microscopy, carbon grids with a 400-mesh size (Electron Microscopy Sciences, CF-400-Cu-50) were subjected to glow discharge using a PELCO easiGlow Discharge Cleaning System for 25 s. Then, 5 µl of the purified fibril sample was applied to the grid and left to incubate for 1 min before being removed with filter paper. The grid was then stained with 5 µl of filtered aqueous 2% uranyl acetate solution (Electron Microscopy Sciences, catalogue no. 22400) for 1 min; excess stain was absorbed using filter paper. After drying, the grid was imaged using a JEM-1400 Plus transmission electron microscope, equipped with a LaB6 source operating at 120 kV and an AMT-BioSprint 16M CCD camera. For scanning electron microscopy, samples were fixed with 2.5% (v/v) glutaraldehyde in 0.1 M sodium cacodylate buffer overnight at 4 °C. After three rinses in 0.1 M sodium cacodylate buffer, they were postfixed with 2% osmium tetroxide in 0.1 M sodium cacodylate buffer for 2 h. The samples were then rinsed with water and dehydrated with increasing concentrations of ethanol, followed by increasing concentrations of hexamethyldisilazane in ethanol. Cells on coverslips were air-dried under a hood, mounted on scanning electron microscopy stubs with carbon tape, and sputter-coated with gold/palladium using a Cressington 108 auto sputter coater. Images were acquired with a field-emission scanning electron microscope (Zeiss Sigma) at an accelerating voltage of 3 kV and a 15-degree tilt.
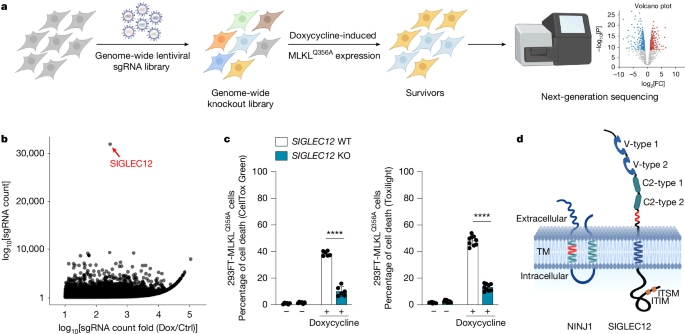
Genome-wide CRISPR-based knockout screens
Human Brunello sgRNA library (Addgene, catalogue no. 73178-LV) was used40. This library contains 76,441 sgRNAs targeting 19,114 genes (4 sgRNAs per gene), with 1,000 non-targeting controls40. HEK293-FlpIn-TREx-MLKLQ356A cells were transduced at a multiplicity of infection of 0.5 using 8 µg ml−1 polybrene, and the plates were spun at 1,000g for 60 min at room temperature (spinfection). Two days later, cells were expanded and selected with 2 µg ml−1 puromycin for 2 weeks. The knockout library was treated with 0.1 µg ml−1 doxycycline to induce MLKLQ356A expression and necroptotic cell death for 3 days (with daily feeding for 3 days to remove dead cells). Genomic DNA from the surviving cells was isolated (using Tissue/Blood DNeasy kit from Qiagen; catalogue no. 69504), and PCR was performed using Emerald Taq (Takara Bio, catalogue no. RR310B) with P7 and P5 primers to amplify the sgRNA sequences, which were then subjected to next-generation sequencing on an Illumina NextSeq 500 with a read configuration of 100 bp, single-end. All the fastq files underwent routine quality checks using FastQC (v.0.11.2; http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and FastQ Screen (v.0.4.4; http://www.bioinformatics.babraham.ac.uk/projects/fastq_screen). The trimmed fastq files were mapped to the reference sgRNA library with a mismatch option set to 0 using MAGeCK. Read counts for each sgRNA were generated, and median normalization was performed to adjust for library sizes. Positively and negatively selected sgRNAs and genes were identified using the default parameters of MAGeCK.
Quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Cells were lysed directly in TRIzol, and RNA was purified by chloroform phase separation and isopropanol precipitation. The RNA pellet was washed with 75% ethanol, air-dried and resuspended in RNase-free water. RNA concentration and purity were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized from 1 μg total RNA using EcoDry Premix with Random Hexamers (Takara Bio, catalogue no. 639547). The RNA was added directly to the lyophilized premix, incubated at 42 °C for 1 h and heat-inactivated at 85 °C for 5 min. Quantitative PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) on a QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific). Each reaction was run in triplicate with the following cycling conditions: 95 °C for 2 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 30 s and 72 °C for 30 s. Gene expression was normalized to that of ACTB using the ΔΔCt method, and data were expressed as fold change relative to control samples. Expression of inflammatory cytokines and chemokines was assessed using gene-specific primers as follows: CXCL1 (forward 5′-AGGGAATTCACCCCAAGAAC-3′, reverse 5′-TGGATTTGTCACTGTTCAGCA-3′); TNFA (forward 5′-CAGAGGGCCTGTACCTCATC-3′, reverse 5′-GGAAGACCCCTCCCAGATAG-3′); IL1B (forward 5′-AAGTACCTGAGCTCGCCAGTGA-3′, reverse 5′-TGCTGTAGTGGTGGTCGGAGAT-3′); CXCL8 (forward 5′-TCTGCAGCTCTGTGTGAAGG-3′, reverse 5′-AATTTCTGTGTTGGCGCAGT-3′).
Proteomics
Samples were digested overnight with trypsin (Pierce) following reduction and alkylation with DTT and iodoacetamide (Sigma-Aldrich). Following solid-phase extraction cleanup with an Oasis HLB µElution Plate (Waters), the resulting peptides were reconstituted in 10 µl of 2% (v/v) acetonitrile (ACN) and 0.1% trifluoroacetic acid in water. Then, 2 µl of each sample was injected into an Orbitrap Fusion Lumos mass spectrometer (Thermo Electron) coupled to an Ultimate 3000 RSLC-Nano liquid chromatography system (Dionex). Samples were injected into a 75 μm i.d., 75-cm-long EasySpray column (Thermo) and eluted with a gradient from 0–28% buffer B over 90 min. Buffer A contained 2% (v/v) ACN and 0.1% formic acid in water, and buffer B contained 80% (v/v) ACN, 10% (v/v) trifluoroethanol and 0.1% formic acid in water. The mass spectrometer was operated in positive ion mode with a source voltage of 1.5 kV and an ion transfer tube temperature of 300 °C. Mass spectrometry scans were acquired at 120,000 resolution in the Orbitrap, and up to ten tandem mass spectra spectra were obtained in the Orbitrap for each full spectrum acquired using higher-energy collisional dissociation for ions with charges 2–7. Dynamic exclusion was set for 25 s after an ion had been selected for fragmentation. Raw mass spectrometry data files were analysed using Proteome Discoverer v.2.4 SP1 (Thermo), with peptide identification performed using Sequest HT searching against the human reviewed protein database from UniProt41. Fragment and precursor tolerances of 10 ppm and 0.6 Da were specified, and three missed cleavages were allowed. Carbamidomethylation of Cys was set as a fixed modification, and oxidation of Met was set as a variable modification. The false discovery rate cutoff was 1% for all peptides.
Statistical and bioinformatics analysis
For all experiments, unless otherwise indicated, n was at least 3. Statistical analyses were performed using Prism (GraphPad Software). Data were analysed using one-way analysis of variance with Bonferroni post-test. Student’s t-test was used for paired datasets. Data points indicate the mean ± s.d. Alignments were done using Clustal Omega42 and visualized using Jalview43. Secondary structure predictions were obtained from UniProt41.
ProteinAtlas data analysis
The clustering data for the gene expression sets from the deep sequencing of RNA from 40 different normal tissue types (https://www.proteinatlas.org/ENSG00000254521-SIGLEC12/tissue) and scRNA-seq clustering data from 31 human tissue types (https://www.proteinatlas.org/ENSG00000254521-SIGLEC12/single+cell) were obtained from the ProteinAtlas database (v.22)44. UMAP plots display gene clusters from Louvain clustering of gene expression across all tissue types or single cell types.
Human mutation analyses
To identify the most prevalent SIGLEC12 point mutations in the general population, we conducted a comprehensive analysis using the dbSNP database. We specifically targeted missense mutations owing to their potential impact on protein function. The search involved filtering for missense mutations in the specified genes and extracting the relevant data. We isolated the necessary columns and ranked the mutations in descending order on the basis of allele frequency aggregator (ALFA) values. The ALFA value represents the aggregation of allele frequency across diverse populations, providing a comprehensive measure of mutation prevalence. From this analysis, we identified the top ten mutations with the highest ALFA values, implicating the most significant missense mutations in the general population. To identify the most frequent SIGLEC12 mutations associated with cancer, TCGA was analysed using cBioportal45.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.