Mice
Female and male C57BL/6J (Jackson Laboratory, 664) and heterozygous transgenic TRAP2;Ai14 (ref. 50), Amigo2-Cre2 (B6.Cg-Tg(Amigo2-cre)1Sieg/J; Jackson Laboratory, 30215), Sert-Cre (Mouse Mutant Resource and Research Centers, stock number 017260-UCD, strain code: Tg(Slc6a4-cre)ET33Gsat/Mmucd), NT-Cre44 (a gift from S. Russo) and, Shank3Δ4-22 (ref. 39) (B6.Cg-Shank3tm2.1Bux/J; Jackson Laboratory, 32169), dCas9-KRAB (Rosa26-LSL-dCas9-KRAB (Jackson Laboratory, 033066) mice were used as experimental subjects. TRAP2;Ai14 mice were generated by crossing TRAP2 (STOCK Fostm2.1(icre/ERT2)Luo/J; Jackson Laboratory, 30323) and Ai14 (B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J; Jackson Laboratory, 7914) mice. Male retired breeder CD-1 (Charles River, ICR), female Swiss-Webster (Charles River, CFW) as well as male and female C57BL/6J mice were used as partner mice.
Except for singly housed aggressors, mice were housed with 2–5 mice per cage and weaned at 21 days old. All behavioural experiments were conducted with 7–14-week-old mice. Animals were maintained on a 12-h light–dark cycle at roughly 21 °C with 50% humidity and food and water ad libitum. Behavioural experiments were performed during the same circadian period (07:00–19:00). Experiments were conducted in accordance with the National Institutes of Health Guide for Care and approved by the Use of Laboratory Animals and the Icahn School of Medicine at Mount Sinai Institutional Animal Care and Use Committee. Sample sizes were not predetermined by statistical analysis but based on previous experience with the variance of the assays. The sequence of control and manipulated conditions were shuffled.
Stereotactic injections and cannula implantation
Mice (4–8 weeks old) were anaesthetized with intramuscular injections of a drug mixture containing ketamine (100 mg kg−1) and xylazine (5 mg kg−1) and heads were then positioned on a stereotaxic apparatus (David Kopf Instruments) for virus injections (0.2–0.5 μl) at a flow rate of 0.1–0.25 μl min−1 with a microinjection pump (Harvard Apparatus). The following bregma coordinates from dura were used to target the virus solution filled glass cannula: dCA2 (bilateral, anteroposterior −1.6 mm; mediolateral ±1.6 mm; dorsoventral 1.7 mm), vCA1 (bilateral, anteroposterior −3.16; mediolateral ±3.1; dorsoventral −4.55), DR (unilateral, anteroposterior −4.36; mediolateral 0; dorsoventral −3.1), aPVT (unilateral, anteroposterior −0.4; mediolateral 0; dorsoventral −3.8), median raphe (unilateral, anteroposterior −4.4; mediolateral 0; dorsoventral −4) and reuniens (bilateral, anteroposterior −0.2; mediolateral ±0.85; dorsoventral −4.3).
Adeno-associated viruses (AAVs) used for stereotaxic injections were purchased from Addgene, WZ Biosciences and the Stanford Neuroscience Gene Vector and Virus Core and included: AAVDJ-hSyn-EGFP, AAVDJ-hSyn-hM4Di-mCh, AAVDJ-hSyn-DIO-hM4D(Gi)-mCh, AAVDJ-hSyn-mCh, AAVDJ-EF1α-DIO-eYFP, AAVDJ-hSyn-ChR2-eYFP, AAVDJ-hSyn-ChR2-mCh, AAVDJ-hSyn-GCaMP6f, AAVDJ-EF1α-DIO-RCaMP2, AAVDJ-EF1α-DIO-NpHR3.0eYFP, AAVDJ-EF1α-DIO-ChR2-eYFP, AAVDJ-EF1α-DIO-ChR2-mCh, AAVDJ-CMV-DIO-EGFP, AAVDJ-EF1α-DIO-mCh, AAVDJ-EF1α mCh-IRES-Cre-WPRE, AAV9-hSyn-EGFP-CAAX, AAV9-hSyn-GRAB5-HT3.5 and AAV9-hSyn-GRABNT1.0. AAV titres ranged from 1 × 1012 to 2 × 1013 gc ml−1. CAV2-Cre (ref. 51) was purchased from Plateforme de Vectorologie de Montpellier. Synapsin-driven lentiviral NLS-GFP-Cre, AAV-CAG-FLEx-TC, AAV-CAG-FLEx-G and rabies virus (Janelia Research Campus) were used for monosynaptic tracing. Behavioural experiments with cell body manipulations were performed 3–4 weeks after virus injections, whereas the viral particles were allowed to incubate 4–8 weeks for manipulation of axon terminals.
Optic fibres (400-µm core, numerical aperture (NA) 0.5; RWD Life Science) for optogenetic light delivery were implanted in the vCA1 bilaterally at anteroposterior −3.16; mediolateral ±3.1; dorsoventral −4.4. A 26-gauge guide cannula (Plastics One) for drug micro-infusion was implanted at anteroposterior −3.16; mediolateral ±3.1, so that the 33-gauge infuser insert reached a depth of dorsoventral −4.5. Miniature screws (thread size 00–90 × 1/16; Antrin Miniature Specialties and McMaster-Carr), light-cured dental adhesive cement (C&B Metabond, Parkell) and resin (Geristore A&B paste, DenMat or Ortho-Jet powder&liquid, Lang Dental) were applied to secure implants to the skull. A small percentage (roughly 5%) of mice were excluded from behavioural analysis based on either off-target transgene expression or inaccurate implant placement.
Intraperitoneal injection and cannula infusion of drugs
For this, 4-hydroxytamoxifen (4-OHT, Sigma H6278-50MG) was administered by means of intraperitoneal (i.p.) injection at 50 mg kg−1 for 15 min after pairing with partner mice. CNO (Tocris Biosciences 4936) was also delivered through i.p. injections at 10 mg kg−1 for 30 min before behavioural experiments. Drugs used for cannula microinjection experiments were infused in a total volume of 200–400 nl at a speed of 200 nl min−1 through an injector cannula connected to a micro-infusion pump (World Precision Instruments). The following drugs and concentrations were used: methiothepin mesylate salt (Millipore Sigma M149, 0.2 μg), NAS-181 (Tocris Biosciences 1413, 2 μg), NAD-229 (Tocris Biosciences 3282, 3.5 μg), SR48692 (Tocris Biosciences 3721, 5 pg), NTRC 824 (Tocris Biosciences 5438, 22 pg) and CP93129 dihydrochloride (Tocris Biosciences 1032, 0.5 μg). After completion of the infusion, injection infusers stayed for 2 min until removal and sociability and social memory assays were carried out 20 min later. NAS-181 (10 mg kg−1) and SR48692 (3 mg kg−1) were delivered through i.p. injections 30 min before fibre photometry experiments.
TRAP2;Ai14 behavioural experiments
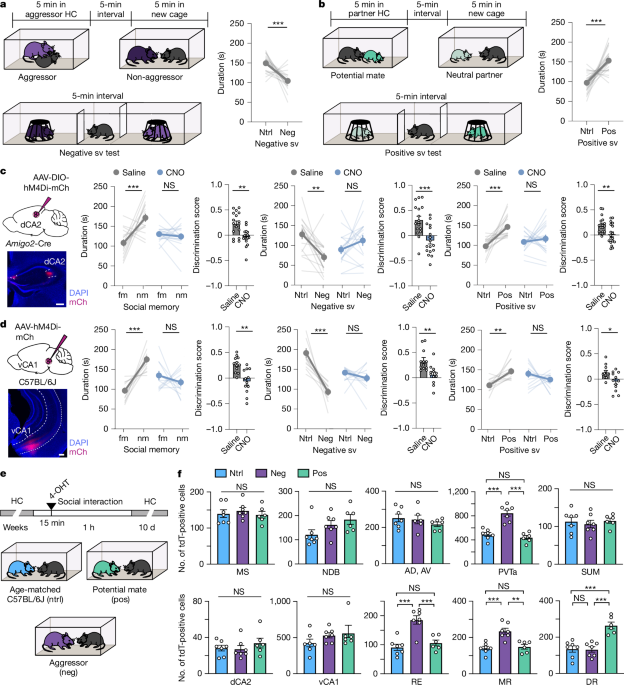
Three groups of TRAP2;Ai14 mice (6–8 weeks old) were habituated to i.p. saline injections and empty cages for two consecutive days. On test day, group 1 subjects were introduced to an age-matched same-sex conspecific in a new cage, group 2 to a sex-matched aggressive male CD-1 or female CFW mouse in the aggressor’s home cage and group 3 to a potential mate (opposite sex) in either the female partner’s home cage or the female subject’s home cage. After 15 min of interaction, subjects were administered 4-OHT (50 mg kg−1) and interaction was continued for 1 h before subjects returned to their home cages. Ten days later, mice were either perfused with 10% neutral buffered formalin (NBF) for histological analysis or brains were removed to be fresh frozen for in situ hybridization experiments.
Three-chamber behavioural tests
Depending on the experiment, test mice were habituated for 2 days to i.p. injections, infuser inserts or the fibre optic patch cord as well as the three-chamber apparatus containing two empty inverted metal grid pencil cups (10 cm diameter) in the outer two chamber for 5 min. The three-chamber apparatus (60 cm long by 23 cm wide by 26 cm high) was constructed of 0.3-cm-thick white opaque acrylic sheets, with two outer chambers (23 cm long by 23 cm wide) and a centre chamber (15 cm long by 23 cm wide) divided by 15-cm-long barriers extending from opposite ends of the walls. White opaque acrylic dividers were used to block entrance to the two outer chambers from the centre chamber. Partner mice were also habituated under the cup for 5 min for 2 days. On test day, subjects were placed into the centre chamber for 2 min before the barriers were lifted, whereafter the subjects were able to freely explore all three chambers. Four variations of this assay were performed to test social memory. The location of partner mice and objects in either chamber was counterbalanced between trials in all tests.
In the neutral social memory test, the first 10-min session is also a sociability test, in which the time spent in the chamber with a same-sex age-matched novel mouse or an object is measured similarly to previously described methods3,5,7. After a 10-min interval, in which subjects were separated from the contents of the two outer chambers, the barriers were lifted and the subjects were allowed to freely explore the three chambers for 5 min in a second session. This time, the one of the outer two chambers contained a novel mouse and the other one a familiar mouse (same mouse from previous session) under a cup.
In the negative social memory test, subjects interacted directly or 5 min with either a same-sex aggressor in the aggressor’s home cage (similar to a resident–intruder test17,18) or a same-sex non-aggressor in a new cage. After a 5-min interval, those subjects that interacted with an aggressor were exposed to a non-aggressor for 5 min and vice versa. The sequence was counterbalanced between trials. After another 5-min interval, the subject was placed into the centre chamber for 2 min, whereafter the barriers were lifted and the subject was allowed to freely explore the three chambers containing the aggressor and the non-aggressor under a cup for 5 min. Owing to the sharp drop in the discrimination scores after a short separation time3,7, a 5-min interval was chosen to shorten the separation time after the first interaction. Singly housed (at least 10 days) male retired breeder CD-1s were used as male aggressors and pair-housed female CFWs were used as female aggressors18. CD-1s and CFWs were screened for their level of aggression as previously described17 and only mice that attacked within 10 s and more than three times within 5 min qualified as aggressors. Retired breeder CD-1s and female CFW group-housed with same-sex conspecifics were used as non-aggressors. Only mice that did not attack in a 5-min interaction qualified as non-aggressors.
In the positive social memory test, subjects interacted directly with either an opposite-sex conspecific potential mate in the potential mate’s home cage or an opposite-sex conspecific neutral partner in a new cage for 5 min. After a 5-min interval, the subject that interacted with one type of partner was exposed to other for 5 min, whereby the sequence was counterbalanced between trials. After another 5-min interval, social memory was tested in a three-chamber apparatus for 5 min with one outer chamber containing the potential mate and the other containing the neutral partner under the cup. In the case of male subjects, the experimenter held the potential mate female partner to simulate the mating posture19 with its rear close to the male’s snout three times for 3 s. In the case of female subjects, a barrier was placed into their home cage so the timing and pacing of the interaction with the potential mate male partner could be controlled. The cardboard barrier had an opening at the bottom, small enough for the female, but not the male, to easily escape to the other side of the cage. The interactions with the neutral partner were without lordosis posture and barrier in a new cage. It is important to note that the term neutral is used to describe a more neutral or less positive stimulus compared to the potential mate but not an absolute neutral stimulus.
We used two variations of the RTPP test. The basic RTPP was performed as previously described31, in which light stimulation was paired with one of the two outer chambers while the subject was allowed to freely explore the three chambers for 15 min. Next, light stimulation was paired with the opposite outer chamber while the subject freely explored all three chambers again for 15 min. In variation 1, the two outer chambers each contained a new mouse and light stimulation was paired with one of the two chambers, while the subjects freely explored all three chambers for 10 min. After a 10-min interval, the barriers were lifted and the subjects were allowed to move freely between the three chambers, where the outer chambers contained the two previously acquainted mice (now familiar mice) under a cup. During this 5-min period, no light stimulation was applied. In variation 2 of this assay, two new objects instead of mice were used.
Recorded videos in the three-chamber apparatus was analysed using a video tracking system (BIOBSERVE, v.3.01), which automatically tracked the location of the subject mouse. The time spent in either of the two outer chambers were scored for the duration of the tests. The following were used to calculate the discrimination scores: sociability test ((time in novel mouse chamber − time in object chamber)/(time in novel mouse chamber + time in object chamber)); neutral social memory test ((time in novel mouse chamber − time in familiar mouse chamber)/(time in novel mouse chamber + time in familiar mouse chamber)); negative social memory test ((time in non-aggressor chamber − time in aggressor chamber)/(time in non-aggressor chamber + time in aggressor chamber)); positive social memory ((time in potential mate chamber − time in neutral mouse chamber)/(time in potential mate chamber + time in neutral mouse chamber)); RTPP mouse/object ((time in previously light-paired mouse/object chamber − time in no-light mouse/object chamber)/(time in light-paired mouse/object chamber + time in no-light mouse/object chamber)). For all experiments, male and female subject mice were used in roughly equal numbers. Test mice were excluded from the analysis (less than 2% of the total), if they were not attacked by the aggressor, if the non-aggressor attacked more than three times in 5 min, or if they spent the entirety of the assay in one chamber only.
Optogenetic stimulation
Mice were habituated for 2 days to behavioural chambers with optic fibres connected to light-emitting diodes (LEDs) in off states. A 545-nm Dual-LED (Prizmatix) was connected to the optical implants via a rotary joint and a fibre optic patch cord (Prizmatix) to photostimulate NpHR3.0. A 450-nm Dual-LED (Prizmatix) was used to photostimulate ChR2. The LEDs were adjusted to around 15 mW for axon terminal stimulation using a digital power meter console (ThorLabs) and the frequency was controlled by Pulser Plus (Prizmatix). A cycle of 8 s on and 2 s off was used to stimulate NpHR3.0 to avoid tissue overheating. ChR2 was stimulated at 20 Hz with a 5-ms pulse duration.
CPP
Similar to previously described52, mice were tested for baseline preference for two differently textured clear acrylic floors by spending 15 min in a white opaque polycarbonate chamber (23 cm long by 23 cm wide by 26 cm high, 0.3 cm thick) on both floors (23 cm long by 13 cm wide by 2.5 cm high, 0.3 cm thick). The ‘grid’ floor was constructed of seven evenly spaced apart rails, which were 2.5 cm in width. The hole floor contained 4 × 9 round holes 8 mm in diameter, evenly spaced apart. On day 1, mice were either administered saline or CNO (10 mg kg−1) through i.p. injections 30 min before conditioning to one of the floors for 15 min. On the following day, mice that received saline on the previous day, received CNO and vice versa. 30 min later, animals were conditioned to the alternate flooring for 15 min. On day 3, test mice were placed into the chamber containing both types of flooring and time spent on either type was automatically scored with video tracking software (BIOBSERVE).
Elevated-plus maze
Mice were placed into the centre of the elevated-plus maze (74 cm long by 74 cm wide by 94 cm high; Med Associates Inc.) with white floors (each arm 35 cm long by 6 cm wide with 19 cm high walls on closed arms) for 10 min and location of the mice were scored with video tracking software (BIOBSERVE).
Male and female sexual behavioural tests
Male subjects were placed for 5 min into a female conspecific’s home cage with one female present. The female was held by the experimenter for 3 s in a mating posture, so that the male could sniff the female’s rear. Male mounting attempts during the 5 min of free interaction were manually quantified. As females do not show mounting behaviour, a different test was used to assess female receptiveness to males. Male partners were placed into the female subject’s home cage, which was divided into two compartments by a barrier, small enough for the female to easily escape. Escape times and duration of the female in each compartment were manually scored.
Fibre photometry
AAVDJ-CaMKII-GCaMP6f, AAV9-hSyn-EGFP-CAAX, AAV9-hSyn-GRAB5-HT3.5 (ref. 53) or AAV9-hSyn-GRABNT1.0 (ref. 9) was injected into the vCA1 (unilateral, anteroposterior −3.16; mediolateral ±3.1; dorsoventral −4.55), and fibre optic implants were secured above the injection site (unilateral, anteroposterior −3.16; mediolateral ±3.1; dorsoventral −4.4) in stereotaxic surgeries. Two to three weeks later, mice were habituated to the behavioural set-up and tested on the following day with simultaneous video and fibre photometry acquisition. Test mice interacted with a new object, a novel same-sex age-matched conspecific, a sex-matched aggressor or a potential mate for 5 min with an interval of 5 min, whereby the sequence of the interaction was counterbalanced between subjects. The interaction with the aggressor and female potential mate was in the partner’s home cage, whereas the interaction with the object and same-sex conspecific was in a new cage. Female subjects interacted with the potential mate in their own home cage. Synapse software, which controlled an RZ10X LUX-I/O processor (Tucker-Davis Technologies), was used for data acquisition. To stimulate Ca2+-dependent and isosbestic emission, GCaMP6f was excited by frequency-modulated built-in 465- and 405-nm LEDs (RZ10X LUX-I/O processor), respectively. All optical signals were band-pass-filtered with a fluorescence mini cube (Doric), emission was measured with built-in photosensors in the RZ10X LUX-I/O processor and the signal was digitized at 6 kHz. A previously described custom MATLAB_R2022b (MathWorks) code7 was used for signal processing. To debleach, fibre photometry raw signals were fitted with a mono- or bi-exponential decay function, and the resulting fluorescence trace was z scored. Corresponding videos were manually analysed by frame in MATLAB to identify the time of physical contact between test mouse and partner mouse and object. Peristimulus time histograms were constructed by averaging 7 s of non-overlapping epochs from the z scored trace, where a time of 0 represents the time of contact. The maximal z score between 0 and 4 s was used as peak z scored fluorescence.
Molecular cloning and AAV production
We generated a multi-guide CRISPRi AAV vector starting with pX552 (Addgene, 60958) as the backbone, which we then digested with NotI-HF (NEB). Between the NotI sites, we inserted two gene fragments to generate the AAV backbone pAAV mU6-sgRNA-CR1 EF1α-EGFP-W3-SV40. The first fragment comprised a mouse U6 (mU6) promoter, multiple cloning site for protospacers, constant region (CR1) and an EF1α promoter derived from pU6-sgRNA EF1α-puro-T2A-BFP (Addgene, 60955) but engineered to include BbsI sites flanking the mU6-sgRNA-CR1 cassette. The second fragment contained EGFP, W3 terminator and SV40 poly(A) signal. Both were assembled into the NotI-digested pX552 backbone using a three-part Gibson assembly (HiFi NEB). Subsequently, sgRNA arrays comprising a mU6 promoter, sgRNA protospacer sequence of interest and CR1 cassette, synthesized as gene fragments, were inserted into the backbone with a Golden gate approach using the engineered BbsI (NEB) sites in accordance with the manufacturer’s instructions. To ensure maximum knockdown efficiency, the vectors consist of three tandem sgRNA cassettes containing the protospacers predicted to have highest activity according to the Weissman laboratory’s V2 CRISPR sgRNA algorithm54.
AAV particles were produced using a previously described protocol with minor modifications55. One T175 flask of HEK293T cells (American Type Culture Collection) per construct was transfected with 87.6 µg PEI (1 µg ml−1; 3:1 PEI: DNA; PolySciences 24765) along with 15.4 µg of pAdΔF6, 7.3 µg of pAAV2/9n (pAdΔF6 and pAAV2/9n were gifts from J. M. Wilson; Addgene, catalogue nos. 112867 and 112865, respectively) and 6.5 µg of pAAV mU6-sgRNA-CR EF1α-EGFP-W3-SV40 plasmids. Viral supernatant was harvested 72 h posttransfection, along with cells and mixed thoroughly with 0.1 volumes of chloroform. NaCl was added to a final concentration of 1 M, mixed thoroughly, and the sample centrifuged for 5 mins at 3,000g, 4 °C. The aqueous phase was retained, mixed with 9.4 ml of 50% PEG-8000, incubated on ice for 1 h and centrifuged for 30 min at 3,000g, 4 °C. Pellets we resuspended in dPBS, mixed with 5 µl benzonase (EMD Millipore, 71205) and 1 mM MgCl2, and incubated at 37 °C for 30 min. Postbenzonase treatment, samples were mixed thoroughly with an equal volume of chloroform and centrifuged for 5 min at 3,000g, 4 °C. Samples were buffer exchanged into dPBS and concentrated to roughly 50 µl using 100-kDa cut-off cellulose centrifugal filters (Millipore Sigma, UFC8100).
Immunohistochemistry
For the cFos immunohistochemistry experiment, mice were subjected to an interaction with an aggressor (CD-1/CFW) or a potential mate for 1.5 h and immediately perfused with chilled 10% NBF. After overnight incubation in 10% NBF, 40-μm slices were prepared on a Leica VT 1000S vibratome and collected in 24-well plates filled with phosphate buffered saline (PBS). After a 1.5-h blocking in 5% normal goat serum, slices were incubated with 1:1,000 diluted primary antibody and 5% normal goat serum at 4 °C for 12 h. After three 5-min washes with PBS, slices were incubated for 1.5 h in secondary antibody, subsequently washed three times in PBS for 15 min and Fluoromount-G mounting medium was used for 4,6-diamidino-2-phenylindole (DAPI) staining. The following primary antibodies were used: 1:1,000 diluted cFos antibody (Synaptic Systems, 226 008) and 1:500 diluted TPH2 antibody (Abcam, ab184505). The following secondary antibodies were used: Alexa 488 (1:500 dilution), Alexa 568 (1:1,000 dilution) and Alexa 633 (1:1,000 dilution).
Validation of knockdown efficiency
AAVDJ-EF1α mCh-IRES-Cre-WPRE and AAV9-EF1α-sgTph2-EGFP or AAV9-EF1α-sgNTC-EGFP were injected into the DR (unilateral, anteroposterior −4.36; mediolateral 0; dorsoventral −3.1) of dCas9-KRAB mice. Three weeks later, immunohistochemistry was performed as described above. Imaging was conducted on a Zeiss LSM 780 confocal (×20 objective; NA 0.8; diode laser 405 nm, argon laser 458/488/514 nm, diode-pumped solid-state laser 561 nm and HeNe laser 594/633 nm; detectors 32 channel GaAsP, T-PMT (transmitted light), 2-PMT and Airyscan detector; bit depth of images was 8 bit). TPH2 channels were background subtracted, and the same cut-off threshold was applied for all replicates. Polygons were manually drawn around the DR area and particles within the area of interest were analysed. All infected DR cells received sgTph2 or non-targeting guides. To evaluate cell body TPH2 expression in the DR, we analysed fluorescence intensity. An average percentage fluorescence intensity was calculated from the sgNTC replicates and the remaining ratio was used for calculation of the percentage TPH2 fluorescence intensity and knockdown efficiency.
Behavioural cohorts were injected with AAV9-EF1α-sgTph2-EGFP or AAV9-EF1α-sgNTC-EGFP into the DR and CAV2-Cre into the vCA1. Six weeks later, behavioural experiments were performed followed by perfusion and immunohistochemistry. Imaging and background subtraction was performed as described above. Polygons were manually drawn around the vCA1 and particles in within the area were analysed. In the vCA1 only the DR terminals received sgTph2 or non-targeting sgRNA. Therefore, we analysed the area of the TPH2+ terminals in the vCA1. An average percentage of TPH2+ area within the vCA1 area was calculated from the sgNTC replicates and the remaining ratio was used for calculation of percentage TPH2+ area.
Primary neuronal cultures
Primary neuronal culture was performed as previously described56. In brief, DR nuclei were dissected from P0 pups in ice-cold Hank’s buffered saline solution (HBSS) buffer (Thermo Fisher Scientific, 88284) supplemented with 2 mM Ca2+ and 0.5 mM EGTA under a dissection hood and incubated in Neuronal Isolation Enzyme (Thermo Fisher Scientific, 88285) and HBSS for 20 min, washed twice with HBSS and dissociated in prewarmed Neurobasal-A Medium (Thermo Fisher Scientific, 10888022) with 2% FBS (Avantor, 97068-085) by gentle trituration. Cell yield and viability were determined by a hemocytometer and trypan blue (Thermo Fisher Scientific, 15250061) staining, respectively. Neurons were plated on a poly-d-lysine (Thermo Fisher Scientific, A3890401) precoated 24-well plate. After 24 h, half of the serum medium was replaced with an equivalent volume of Serum-free Neurobasal-A Medium supplemented with 0.5 mM l-glutamine (Thermo Fisher Scientific, A2916801) and 0.5% B-27 (Thermo Fisher Scientific, 17504044). On day 3, half of the old medium was replaced with fresh medium supplemented with 4 mM Ara-C (Sigma, C6645), and neurons were infected with AAV constructs expressing control sgRNAs or Tph2 sgRNAs. After infections, the primary neuronal cultures were maintained for 1 week with medium changes every 3 days.
qPCR with reverse transcription
RNA extraction was performed using a Quick-RNA MicroPrep Kit (Zymo Research, R1050) according to the manufacturer’s protocol. Total RNA was reverse transcribed using the High-Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific, 4387406). The resulting complementary DNA (cDNA) was then used for qPCR using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, A25742) with gene-specific primers. Data were recorded using a QuantStudio 7 Flex Real-Time PCR system (Thermo Fisher Scientific). Tph2 expression was normalized to Actb. The relative changes in expression were calculated using the 2-ΔΔCt method. Primers used in the studies include: Tph2 (mouse): forward, 5′-GCAAGACAGCGGTAGTGTTCT-3′; reverse, 5′-CAGTCCACGAAGATTTCGACTT-3′; Actb (mouse): forward, 5′-GGCTGTATTCCCCTCCATCG-3′; reverse, 5′-CCAGTTGGTAACAATGCCATGT-3′.
Ex vivo electrophysiology
Here, 4–8-week-old C57BL/6J, Shank3Δ4-22+/− and wild-type littermates were used for whole-cell recordings. Mice were euthanized and brains were sliced in a sucrose cutting solution containing (in mM): 228 sucrose, 26 NaHCO3, 11 glucose, 2.5 KCl, 1.2 NaH2PO4, 7 MgCl2 and 0.5 CaCl2 on a vibratome (Leica VT1200 S). Slicing and recording solutions were continuously equilibrated with 95% O2 and 5% CO2. Coronal vCA1 slices (300 μm) were transferred to a slice holding chamber with artificial cerebrospinal fluid containing (in mM): 119 NaCl, 26 NaHCO3, 11 glucose, 2.5 KCl, 1.2 NaH2PO4, 1.3 MgCl2 and 2.5 CaCl2 (osmolarity 289–295) for 30 min at 32 °C and then further equilibrated for 30 min at room temperature. Next, brain slices were placed in a recording chamber perfused with 28–30 °C artificial cerebrospinal fluid and visualized with a ×40 water-immersion objective on an upright fluorescent microscope (BX51WI; Olympus) equipped with infrared-differential interference contrast video microscopy and epifluorescence (CoolLED). Whole-cell current-clamp recordings (pipette opening 4–6 MΩ) were performed with pipettes filled with (in mM): 130 C6H11KO7, 5 KCl, 10 HEPES, 0.6 EGTA, 2.5 MgCl2, 4 Mg2ATP, 0.4 Na3GTP, 10 phosphocreatine (pH 7.25; osmolarity 290). Whole-cell voltage-clamp recordings (pipette opening 3–4 MΩ) of IPSCs were performed with pipettes filled with (in mM): 80 CsCl, 65 CsMeSO4, 8 NaCl, 10 HEPES, 0.25 EGTA, 2 Mg2ATP, 0.3 Na3GTP, 0.1 spermine, 7 phosphocreatine (pH 7.34; osmolarity 300). No CsCl but 140 CsMeSO4 was used for whole-cell voltage-clamp recordings of EPSCs. Series and input resistance were monitored with −4 mV, 70-ms pulse delivered through the recording pipette and experiments were excluded from the analysis if series resistance varied by more than 15%.
Current-clamp recordings were carried out to assess intrinsic cell excitability with a series of incremental rectangular depolarizing current pulses (20 pA, 500 ms) injected into vCA1 pyramidal cells. Next, the same procedure was performed in the same cell after either CP93129 dihydrochloride (Tocris Biosciences 1032, 5 µM) or PD149163 (Sigma, 0.5 µM) was bath applied for 5 min. Following 10 min of wash-out, recordings were carried out in the same cell in presence of the second drug, whereby the sequence of drug application was counterbalanced between cells. Only one cell was recorded per slice. Current-clamp recordings in the PVT was carried out with 20-pA depolarizing current pulses and in the DR with 25-pA depolarizing current pulses. Pipettes were filled with in (mM): 130 C6H11KO7, 10 KCl, 10 HEPES, 0.2 EGTA, 4 Mg2ATP, 0.5 Na3GTP, 10 phosphocreatine. CNO (Tocris Biosciences 4936, 5 µM) was bath applied for 5 min. Evoked EPSCs and IPSCs were recorded from vCA1 cells with a bipolar stimulating electrode (fabricated from platinum and iridium wire) placed near the recording pipette. Baseline evoked and spontaneous EPSCs were recorded in voltage-clamp settings at −70 mV the presence of picrotoxin (50 μM). Baseline evoked and spontaneous IPSCs were recorded at −70 mV in the presence of NBQX (10 μM) and D-AP5 (50 μM). CP93129 dihydrochloride (5 µM) or PD149163 (0.5 µM) were bath applied after an initial baseline recording. Summary graphs of the effects of CP93129 and PD149163 over time in evoked recordings were generated by averaging 1-min bins as a percentage of the averaged 4-min baseline. Cumulative probability graphs were generated from all spontaneous events before and after drug application. Synaptic responses were recorded in the vCA1 8 weeks after stereotaxic injections of AAVDJ-hSyn-ChR2-eYFP into the DR or PVT. To photostimulate ChR2 in vCA1 axons and/or terminals, 470-nm 0.1–5.0-ms light pulses from the pE-300ultra (CoolLED) were delivered by means of a ×40 water-immersion objective to the whole slice. NAS-181 (20 µM) or SR48692 (0.5 µM) were bath applied following an initial baseline recording. Recordings were made using a MultiClamp 700B amplifier (Molecular Devices), digitized at 10 kHz with the Digidata 1320A or 1440A data acquisition system (Molecular Devices) and analysed with Clampfit v.10.7 software (Molecular Devices).
In situ hybridization and quantification
The experiment was performed according to the manufacturer’s manual57 using the RNAscope Multiplex Fluorescent v.2 kit (Advanced Cell Diagnostics). Briefly, brains were fresh frozen on dry ice and stored at −80 °C until 15-μm slices were prepared and collected directly on Superfrost plus microscopy slides using a cryostat. Following fixation in prechilled 4% paraformaldehyde, slices were washed in PBS and dehydrated gradually in 50%, 70% and two times 100% ethanol (5 min each). After a barrier was created with a hydrophobic pen, slides were incubated with roughly five drops of RNAscope hydrogen peroxide for 10 min, subsequently washed with distilled water and incubated with Protease IV for 30 min, then washed twice with PBS. The probes mCh-O3 (513201), Mm-Slc17a6-C2 (319171-C2), Mm-Gad2-C2 (439371-C2) and Mm-Nts-C3 (420441-C3) were warmed up to 40 °C in a water bath, and 1 volume of C2 and C3 were diluted in 50 volumes of C1 to create the probe mixture. In the hybridization step, slices were incubated with the probe mixture for 2 h at 40 °C in a HyEZ Oven and rinsed twice at room temperature in Wash buffer (50× RNAscope Wash buffer diluted in distilled water). Next, slides were incubated with roughly six drops of RNAscope Multiplex FL v2 Amp1 for 30 min at 40 °C in the HyEZ Oven and washed twice with wash buffer at room temperature. This procedure was repeated with RNAscope Multiplex FL v2 Amp2 (30 min) and RNAscope Multiplex FL v2 Amp3 (15 min). Slides were then incubated with RNAscope Multiplex FL v2 HRP-C1 in the HyEZ Oven at 40 °C for 15 min, rinsed twice in wash buffer at room temperature and incubated in a 1:1,000 diluted Opal 520 dye for 30 min at 40 °C and subsequently rinsed twice in wash buffer. This step was repeated with HRP-C2, HRP-C3 and matching Opal dyes 570 (1:3,000 dilution), 650 (1:1,000 dilution). Finally, Fluoromount-G mounting medium was used for DAPI staining.
Imaging was performed on a Zeiss LSM 780 confocal (×20 objective; NA 0.8; diode laser 405 nm, argon laser 458/488/514 nm, diode-pumped solid-state laser 561 nm and HeNe laser 594/633 nm; detectors 32 channel GaAsP, T-PMT (transmitted light), 2-PMT and Airyscan detector; bit depth of images, 8 bits). A Fiji (ImageJ v.1.52p) macro was used for automated image analysis, which was performed blinded. Based on pixel-intensity threshold, DAPI channels were background subtracted, binarized and converted to masks. Polygons were manually drawn around the vCA1 area and particles in the remaining channels were automatically analysed within the masked areas inside the polygon. The cut-off threshold was visually determined by comparing ascending intensity values with marked location in the images to eliminate background particles.
Statistical methods and reproducibility
For behavioural experiments and analysis, the experimenters were blinded to the virus injection the animals received. Prism10 (GraphPad) was used for statistical analysis. Individual data points are identified by sex in the source data. Parametric statistical tests were only chosen for normally distributed samples. Otherwise, non-parametric statistical tests were performed. One-way analysis of variance (ANOVA) with Tukey’s or Dunnett’s multiple comparison post hoc test was used to determine significance for several treatment comparisons and two-way ANOVA with Tukey’s or Sidak’s multiple comparison post hoc test for several group comparisons and across many time points. P values found in figure legends refer to ANOVA results of the whole group, whereas the asterisks between individual samples represent results of the multiple comparison post hoc tests. A two-tailed paired Student’s t-test was applied for within-group comparison of two treatments and unpaired test for comparison between two groups. When normality was not assumed, the Wilcoxon signed rank test was used for within-group comparison of two treatments, Mann–Whitney test for between group comparison and Kruskal–Wallis with post hoc Dunn’s test for multiple comparisons. All tests are two-sided. NS, not significant. *P < 0.05, **P < 0.01, ***P < 0.001. In all figures, data are shown as mean ± s.e.m.
Representative data are one of: Fig. 1c, 17 mice; Fig. 1d, 12 mice; Fig. 2a, 13 mice; Fig. 2e, 11 mice; Fig. 3a, 12 mice; Fig. 3d, 14 mice; Fig. 3j, 10 mice; Fig. 3l, 10 mice (sgTph2) and 8 mice (sgNTC); Fig. 4a, 12 mice; Fig. 4e, 9 mice; Fig. 4h, 10 cells; Fig. 4i, 11 cells; Fig. 4j, 14 cells (predrug), 11 cells (NTR1 (PD)), 8 cells (5-HT1BR (CP)); Fig. 4k, 6 cells (EPSC), 7 cells (IPSC); Fig. 4l, 8 cells; Fig. 5a, 11 mice; Fig. 5b, 10 mice; Fig. 5h, 10 mice and Fig. 5j, 15 cells (Shank3) and 14 cells (wild-type).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.