Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Mouse embryo recovery
Mice were maintained in accordance with national and international guidelines. All experimentation involving animal subjects was approved by the Institutional Animal Care and Use Committee at Yale School of Medicine and conducted following the approved animal handling protocol (protocol no: 2023-20352). All experimental mice were maintained in specific pathogen-free conditions on a 12âh:12âh light:dark cycle temperature-controlled facility (between 20â26â°C and humidity 30â70%) with free access to water and food, and used from 6 weeks of age. All mice were bred to a mixed CD1 albino background. V. Greco provided the H2B-GFP:Tcf-LEF51 reporter mouse line.
To collect in vivo embryos at gastrulation stage, 5- to 7-week-old female CD1 mice were naturally mated with 12- to 24-week-old male CD1 mice and sacrificed 6.5, 6.75, 7.0 or 7.25 days post coitum. Uteri were recovered and embryos were dissected from deciduae in DMEM medium containing 5% FBS, 20âmM HEPES, and 1à Penicillin-Streptomycin (Gibco) warmed up to 37°. The sex of the embryos was not determined.
Embryo culture at gastrulation
Embryos were cultured in a 1:1 ratio of DMEM F/12 (Gibco) to rat serum (Envigo), with 1à Glutamax (Gibco), 0.2à Penicillin-Streptomycin (Gibco), and 0.2à MEM NEAA (Gibco) in 37â°C at 20% O2 and 5% CO2. Medium was equilibrated in 5% CO2 for â¥15âmin prior to culture.
Tetraploid complementation
Mouse embryos at the pre-implantation 2-cell stage (E1.5) were recovered in KSOM medium by flushing the oviduct, from 5- to 6-week-old CD1 females that were superovulated by injection of 10âIU pregnant mare serum gonadotropin (ProSpec) followed by 10âIU human chorionic gonadotropin (Sigma) after 48âh and were mated with CD1 males. Two-cell-stage embryos were then fused to induce tetraploidy using BTX Embryo Manipulation Electro Cell Fusion System. Fused embryos were transferred to advanced KSOM (Sigma) covered with mineral oil (FUJIFILM) and cultured to blastocyst stage until embryonic stem cell injection. Each host tetraploid blastocyst was injected with 10â15 mouse ES cells, the injected blastocysts were transferred into uterus of the prepared day 2.5 CD1 pseudopregnant surrogates (6 weeks old) which were plugged by vasectomized CD1 male mice. These mouse embryonic stem cell-derived embryos were collected at the certain developmental timeline for subsequent experiments by uterine dissection (as described above).
Metabolism and signal modulation experiments
Chemical inhibitors were administered as follows except where otherwise stated: 2âmM 2-DG52 (Santa Cruz Biotechnology), 20âµM BrPA (Santa Cruz Biotechnology), 10âµM PD0325901 (StemCell Technologies), 10âµM SU5402 (StemCell Technologies), 200âµM galloflavin53 (Cayman Chemical), 100âµM oligomycin54 A (Santa Cruz Biotechnology), 5âµM 6-AM55 (Cayman Chemical), 10âµM YZ98 (Cayman Chemical), 5âµM shikonin56 (Cayman Chemical) and 5âµM azaserine57 (Cayman Chemical or Santa Cruz Biotechnology). Embryos were treated for 4 h, 7âh, 12âh or 18âh. Mesoderm explants were treated for 3âh, 12âh or 16âh. Stem cells were treated for the duration of the experiment as indicated in the figure captions. The proteoglycan sulfation inhibitor NaClO3 (Sigma) was applied at 20âmM. FGF2, FGF4 and FGF8 (all Peprotech) were applied at 50ângâmlâ1.
Nutrient-sparse and rescue experiments
Nutrient-sparse medium was prepared using an Advanced DMEM/F-12 medium devoid of d-glucose, l-serine, l-glutamine and sodium pyruvate (Caisson Laboratories) with the addition of 25âmM sodium bicarbonate (Sigma), 0.2Ã penicillin-streptomycin (Gibco) and 0.2Ã MEM NEAA (Gibco). This was supplemented with the following nutrients, according to each experimental condition: 17âmM glucose (Gibco), 0.5âmM sodium pyruvate (Gibco), and/or 1Ã Glutamax (Gibco). For galactose rescue experiments 20âmM galactose (Sigma) supplemented to the culture medium. For GlcNAc rescue experiments 1âmM or 2âmM GlcNAc (Sigma) supplemented to the culture medium. All embryos were cultured in a 1:1 ratio of customized DMEM and rat serum (Envigo).
Cell culture, gastruloids and in vitro mesoderm-directed-differentiation assay
All mouse embryonic stem cells were cultured at 37â°C in 20% O2 and 5% CO2 and passaged once they had reached 80% confluency. Cells were routinely tested for mycoplasma contamination by PCR. Mouse embryonic stem cells were cultured on gelatinized tissue-cultureâgrade plates in FBS-containing DMEM medium with 2i/LIF (1âμM MEK inhibitor PD0325901, 3âμM GSK-3 inhibitor CHIR99021 and 10ângâmlâ1 LIF). FBS-containing DMEM (Gibco) medium comprised of: 18% inactivated FBS (Gibco), 1.2à penicillin-streptomycin (Gibco), 1.2à Glutamax (Gibco), 1.2à MEM NEAA (Gibco), 1.2âmM sodium pyruvate (Gibco), and 120âµM 2-mercaptoethanol (Gibco). The ERK-KTR mouse embryonic stem cells used in this study were sourced from Simon et al.34 (JAX 035333).
For in vitro mouse gastruloid experiments, we followed the protocol as described20,21. Gastruloids were treated with indicated drugs culture day between 3 and 4 and fixed for subsequent HCR staining or qPCR assay, as described in Dingare et al.7.
For in vitro mesoderm-directed differentiation experiments, we followed the protocol as described58, with some modifications. Cells were passaged (day 0) after reaching 80% confluency into 6-well culture plates or 8-well µ slides (Ibidi) at a density of 20,000 per cm2 in FBS-containing DMEM medium with 10ângâmlâ1 Fgf2 (R&D Systems). Cells were treated at day 1 with 10ângâmlâ1 FGF2, at day 2 with 10ângâmlâ1 FGF2 and 5âµM CHIR99021 (StemCell Technologies), and at day 3 and 4 with 5âµM CHIR99021. Drug treatments added to the culture at least 12âh after FGF2 treatment was started on day 1, or on when cells transitioned to mesoderm state on day 3.
In situ mesoderm explant assay
For mesoderm isolation from E7.25 mouse embryos, we followed a modified protocol as described59. Embryos were collected as described above. Extraembryonic tissue proximal to the amnion was removed, and the cup-shaped embryo was transferred to a dissociation medium consisting of 0.5% Trypsin EDTA (Gibco) and 2.5% Pancreatin (Thermo Scientific Chemicals) for 15âmin at 4â°C. Embryos were washed with collection medium (described earlier) and mesoderm tissue was dissected using insect pins (Roboz) attached to syringes. Explants were transferred to fibronectin-coated 18-well µ slides (Ibidi) plates and left to adhere for 3â4âh in medium containing a 1:4 ratio of rat serum to embryo culture medium (described earlier), prior to downstream experiments and/or chemical inhibitor treatments. For live imaging of migration dynamics, explants were imaged every 5âmin up to 3âh. For immunofluorescence staining, explants were cultured up to 17âh and fixed in PBS containing 4% paraformaldehyde (PFA) for 20âmin. For RNA sequencing, explants were cultured to 27âh then scraped off with sterile insect pins and flash-frozen.
Glucose-uptake assay
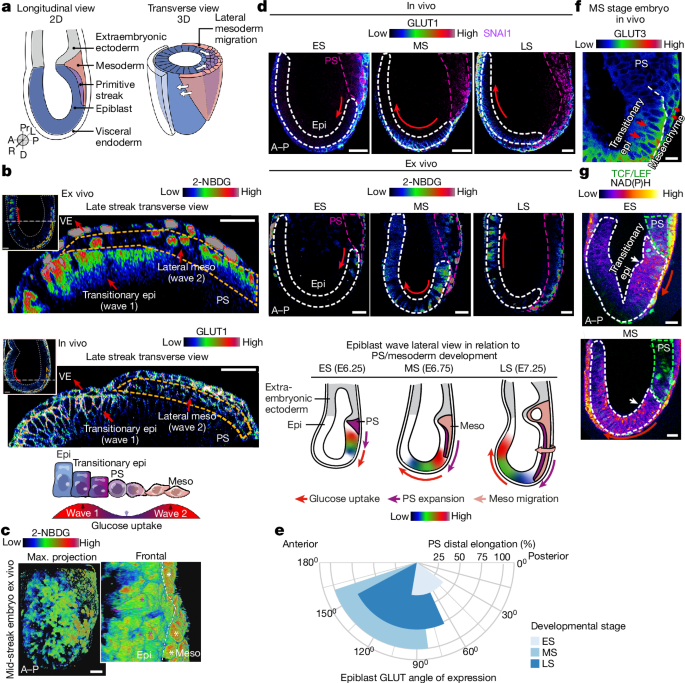
Mouse embryos were collected at early streak, mid streak and late streak stages of gastrulation and cultured with 1âmM 2-NBDG60 (Cayman Chemicals) for 2âh. For multiphoton microscopy, embryos were immediately live-imaged. For confocal microscopy, embryos were fixed in PBS containing 4% PFA for 15âmin then immediately imaged after a 5âmin wash in PBS-T (PBS with 0.05% Tween-20).
DQ gelatin assay
Cells underwent mesoderm-directed differentiation on 8-well µ slides (Ibidi) as previously described. On day 2.5 when cells are at the EpiSC stage, cells were cultured with differentiation medium that included 50âµgâmlâ1 DQ gelatin (Invitrogen), then were fixed on day 3.5 in PBS containing 4% PFA for 20âmin, protected from light. Cells were incubated in blocking buffer (described below) with DAPI (overnight at 4â°C or 20âmin room temperature) prior to imaging.
Invadopodia assay
18-well µ slides (Ibidi) were prepared with FITCâFibronectin (Sigma-Aldrich) and 0.1% gelatin following the manufacturerâs 2-day protocol and protected from light. Explants were cultured on these plates for 16âh prior to fixation in PBS containing 4% PFA for 20âmin. Explants were incubated in blocking buffer (described below) with DAPI (overnight at 4â°C or 20âmin room temperature) prior to imaging.
Labelling active mitochondria
Mouse gastrulas were collected at ES stage and cultured with either 200ânM MitoTracker Deep Red or 100ânM tetramethylrhodamine, methyl ester, perchlorate (TMRM). Embryos were cultured in the respective dyes for 30âmin and then imaged in a humidified chamber with 37â°C and 5% CO2 according to the same parameters described below for live imaging.
Immunofluorescence staining
Samples were fixed in PBS containing 4% PFA for 20-45âmin room temperature, or in methanol for 20âmin at 4â°C where stated. After fixation, samples were washed twice with PBS-T (PBS with 0.05% Tween-20) and permeabilized in PBS with 1âmM glycine and 0.3% Triton X-100 for 20â60âmin at room temperature. Primary antibody incubations took place overnight at 4â°C in blocking buffer (PBS containing 10% fetal bovine serum (FBS), 10% Tween-20). Samples were washed twice with PBS-T prior to secondary antibody incubations at 4â°C in blocking buffer. On the final day, samples were washed twice with PBS-T, then transferred into PBS-A droplets (PBS with 0.75% Bovine Albumin Fraction V (Gibco)) and covered with mineral oil (Sigma-Aldrich) in 35âmm glass-bottom dishes (MatTek) before confocal imaging. All PBS-T washes were done for 10âmin room temperature, and all room temperature incubations or washes took place on a rocking platform. All antibodies used in this study are listed in Supplementary Table 1.
Image data acquisition and processing
Samples were imaged with the Leica STELLARIS 5 microscope using a HC PL APO CS2 40Ã/1.10 or a HC FLUOTAR L 25Ã/0.95âW VISIR 0.17 water objectives, a z-spacing of 0.75âµm to 5âµm and appropriate laser and filters for Alexa 405, Alexa 488, Alexa 546 and Alexa 633 or combinations thereof. To correct for fluorescence decay along the z axis during embryo imaging, âz-compensation by AOTF and PMTâ was defined in a control embryo and applied across all experimental conditions during that imaging session, so that changes in laser power and gain across the z-stack were equivalent across conditions, and only adjusted to each embryoâs size. Raw data were processed using open-source image analysis software Fiji/ImageJ2 2.9.0 or AIVIA 10.5.1 AI Image Analysis Software and assembled in Photoshop 2021 22.3.1 (Adobe) or Illustrator 2024 28.6 (Adobe). Transverse views were generated from 1âµm z-spaced images using ImageJâs orthogonal viewer. Digital quantifications and immunofluorescence signal intensity graphs were obtained using plot profile measurements in Image J and visualized in GraphPad Prism10.2.3. software.
Time-lapse live imaging
Confocal time-lapse imaging of embryos, mesoderm explants, and mesoderm-differentiated cell cultures were performed using Leica STELLARIS 5 microscope using a 25à (HC FLUOTAR L 25Ã/0.95âW VISIR 0.17) or 40à (HC PL APO CS2 40Ã/1.10) water objective and appropriate laser/filters for Alexa 488, Alexa 546 and Alexa 633 or combinations thereof. Samples were imaged under a humidified chamber with 37â°C and 5% CO2. Explants were imaged at 5âmin intervals in 2âµm z-spaced planes for up to 4âh on pre-treated ibidi dishes (Ibidi). Images were processed using AIVIA, described below under âImage analysisâ.
Multiphoton live imaging for NAD(P)H autofluorescence
Multicolour two-photon microscopy was used for live-embryo imaging of NAD(P)H dynamics. Embryos kept in ex vivo culture medium (described earlier) with the addition of 20âmM HEPES. Images were acquired with LA Vision TriM Scope II (LaVision Biotec) laser scanning microscope equipped with a Chameleon Vision II and Discovery ultrafast lasers (Coherent) for different wavelengths imaged sequentially after each z-section15. Wavelengths of 750ânm were used for NAD(P)H and 940ânm for H2B-GFP:Tcf-LEF or ERK-KTRmClover reporter. Although excitation ranges of NAD(P)H and GFP overlapped, their emission was separated by band pass filters: blue range (425â475ânm; NAD(P)H) and green range (500â550ânm; FAD). Exclusion of nuclear GFP signal (in H2B-GFP:Tcf-LEF embryos) from NAD(P)H channel validated separation of emission signals of NAD(P)H and GFP. Embryos were imaged using a 40à water immersion lens (Nikon; NA 1.15) at 400âHz with pixel size of 0.2âµm or 0.3âµm and a z-step of 1 or 1.5âµm. For optimal signal-to-noise ratio, Line averaging of 2 was done for all NAD(P)H images. The fluorescence detected from the reduced metabolites through this method captures both NADH and NADPH (thus called NAD(P)H)61. It should be noted that the intracellular concentrations of the non-phosphorylated NADH and NAD+ is much higher than NAD(P)H and NADP+ (ref. 62). We further validated that NAD(P)H fluorescence signal in the embryos closely co-localized and followed 2-NBDG uptake.
Image analysis
All embryos were positioned exactly along the AâP axis (refer to figures) during imaging for easier quantifications.
Angle of GLUT expression in the epiblast
Mid-embryo sagittal sections of embryos stained for GLUT1 and GLUT3 and with DAPI were used for glucose-uptake quantification in Fiji/ImageJ2. For each embryo, the angle vertex was allocated at the proximal-most boundary between epiblast and extraembryonic ectoderm, at the mid-point between posterior-most (0°) and anterior-most (180°) epiblast. Two angle values were calculated for every embryo (âGLUT startâ and âGLUT endâ) to capture the range of observable GLUT expression in the epiblast, along with the primitive streak distal length of the embryo to allocate Theiler staging (ES, MS or LS, as described above). âGLUT startâ and âGLUT endâ means were calculated for each stage and visualized in a rose diagram with RStudio.
Primitive streak distal elongation percentage
Mid-embryo sagittal sections of DAPI-stained embryos were used for primitive streak distal elongation quantification in Fiji/ImageJ2. For each embryo, the 0% cut-off was marked by anterior epiblast morphology, the 100% cut-off was assigned to the distal-most epiblast cell, and âPSâ (primitive streak) was marked at the distal-most point where the primitive streak morphology extends (Fig. 2b). Measurements were obtained for the vertical distance between 0% and 100% and between 0% and âPSâ, so that each embryoâs primitive streak elongation percentage (0-to-PS divided by 0-to-100) is adjusted to its size. Thus, a control embryoâs primitive streak distal elongation percentage of 95% (Fig. 2b) can be interpreted as âthis embryo has elongated its primitive streak to 95% of its epiblast heightâ. Theiler stages could be assigned with this measurement: â¤50 for ES; between 50âandâ100 for MS; >100 for LS.
Basement membrane breakdown
Mid-embryo sagittal sections of laminin and DAPI-stained embryos were used for basement membrane calculations in Fiji/ImageJ2. For each embryo, the 0% cut-off was marked by anterior epiblast morphology, the 100% cut-off was assigned to the distal-most epiblast cell, and âintact BMâ was marked at the distal-most point where laminin was still intact (Fig. 2d). Measurements were obtained for the vertical distance between 0% and 100% and between 0% and âBMâ, so that each embryoâs basement membrane breakdown percentage (0-to-BM divided by 0-to-100) is adjusted to its size. Thus, a control embryoâs basement membrane breakdown readout of 96% (Fig. 2d) can be interpreted as âthis embryo has broken down 96% of its vertical basement membrane lengthâ.
FITCâfibronectin invadopodia assay
z-sections of DAPI-stained mesoderm explants imaged with a 40à objective were used for fibronectin perforation calculations, with â¥3 images captured per explant. For every image, cell number and perforation number were calculated. Invadapodia degradation is represented as a percentage (perforation number divided by cell number).
ERK-KTR nuclear:cytoplasmic ratio quantification
Sagittal sections of ERK-KTR embryos were used to quantify ERK activity. Manual segmentations were drawn for each cell in Fiji/ImageJ2 to delineate nuclear and cytoplasmic areas, using ERK-KTR and brightfield channels to verify cell morphologies (Extended Data Fig. 8b). For every cell, the following measurements were quantified: nuclear area na, cytoplasmic area ca (including the nucleus), nuclear ERK-KTR intensity ni, and nuclear-subtracted cytoplasmic ERK-KTR intensity ci. To calculate the nuclear:cytoplasmic ratio, the average nuclear intensity (ni/na) was divided by the average cytoplasmic intensity (ci/(caâââna)), such that a ratio â¥1 indicates no ERK activity.
Proliferation quantification
Live-imaged videos captured with a 40Ã objective were used for proliferation counts of TCF-LEF reporter mesoderm explants, quantified manually in Fiji/ImageJ2. For each explant, an overall cell count for âstarting populationâ was calculated on the first frame, and a âcell divisionâ event (TCF-LEF telophase observation) was also assigned by careful frame-to-frame assessment over the course of the video. A proliferation index (cell division number/starting population) was then obtained for each explant, such that a highest index of 1 can be interpreted as âevery cell at the beginning of the video has divided by the end of the videoâ. This was then adjusted to the total frame length of the video so that explants across different experimental replicates and video lengths could be compared. Thus, a control explant with a readout of 16.5% (Fig. 4b) can be interpreted as â16.5% of the mesoderm cells at the beginning of the video have divided by the end of the videoâ.
AIVIA-based image analysis
AIVIA 10.5.1 AI image analysis software was used to examine mesoderm explant migration dynamics. Nuclear segmentations were generated using a âcell-trackingâ recipe, applying a pixel classifier that was trained on TCF-LEF nuclear fluorescent channels from videos of each treatment group in every experimental replicate (Fig. 4b). Parameter values were modified between rounds of pixel classifier âpreviewingâ and training to ensure nuclear segmentation accuracy. Every track was manually examined against the brightfield channel to verify detection accuracy. Incorrect lineages were corrected in the âtrack editorâ or discarded from the final dataset, and parent and daughter tracks were treated independently. Every track measurement of interest was exported to Excel and normalized to its detection length (âfirst frameâ subtracted from âlast frameâ) so that lineages of different detection lengths could be compared. Thus, all data points are plotted in µmâminâ1 or µmâsâ1.
Directed-differentiated mesoderm cell counting
Imaris 10.0.1 software was used to count T-positive, total, and pyknotic cells. Nuclear segmentations were generated using the âSurfacesâ function. Parameters such as background subtraction and morphological splitting were adjusted using the Creation Wizard tool to identify the optimal algorithm settings. Pyknotic cells were counted in the DAPI channel using an increased threshold for background subtraction and a smaller diameter for segmentation than when counting healthy cells using the DAPI signal. Surfaces were examined relative to the original staining throughout creation and in their final state to ensure accuracy.
Western blot
Protein was extracted using RIPA buffer (Thermo Scientific 89900). The reagents were supplemented with 1à protease inhibitor (Thermo Fisher Scientific, 87786) and 1à phosphatase inhibitor (Phos Stop Roche 4906845001) to prevent protein degradation and dephosphorylation during extraction. The protein concentrations were quantified using Pierce BCA. protein assay kit (ThermoFisher, 23225) following the manufacturerâs instructions. The protein was denatured at 95â100â°C for 5âmin in 1à NuPAGE LDS sample buffer (ThermoFisher, NP0007) containing 10% v/v β-mercaptoethanol (Millipore Sigma, M6250). The protein electrophoresis was performed using 4â20% precast polyacrylamide gels (Bio-Rad, 4568095) in 1à Tris/Glycine/SDS buffer (diluted from Bio-Rad, 1610732) at 110âV for 1âh for all proteins. The proteins were transferred from the gel onto a nitrocellulose membrane (ThermoFisher, IB301032) by wet transfer for 1âh at 100âV using the Bio-Rad Mini Trans-Blot Cell following the manufacturerâs instructions. Wet transfer buffer was 20% methanol, 200âmM glycine, and 250âmM Tris. The membrane was blocked in 5% milk in TBST for 1âh at room temperature with gentle shaking. Primary antibodies were incubated with the membrane at recommended concentrations at either 1âh room temperature or 4â°C overnight with gentle shaking. The membrane was washed three times in TBST for 5âmin each, followed by incubation with horseradish peroxidase-conjugated secondary antibodies diluted at 1:5,000 ratio in 5% non-fat milk in TBST for 1âh at room temperature. The blots were then washed three times in TBST for 5âmin each. Blots were treated with SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher, 34094) for 2â5âmin at room temperature. The blots were imaged using a CCD camera-based imager ProteinSimple FluorChem E system.
qPCR with reverse transcription
Total RNA was extracted from cells using an RNeasy Micro Kit per the manufacturerâs instructions (Qiagen). cDNA synthesis was performed with 1 μg of total RNA using a High-Capacity cDNA Reverse Transcription Kit according to the manufacturerâs instructions (Applied Biosystems). The amounts of mRNA were measured using the PowerUp SYBR Green PCR Master Mix (Applied Biosystems). Relative levels of transcript expression were assessed by the ÎÎCt method, with Gapdh as an endogenous control. For primers used in qPCR with reverse transcription, see Supplementary Table 2.
RNA-sequencing sample collection and data analysis
Total RNA was extracted from mesoderm explants using a PicoPure RNA Isolation Kit per the manufacturerâs instructions (Applied Biosystems). RNA samples were submitted to the Yale Center for Genome Analysis for quality assessment, library preparation and sequencing. Each RNA sample contained explants from 2 embryos, and 2 samples were submitted per condition. Paired-end reads were aligned using Star (v2.7.9a) to the mouse genome (GRCm38) using the Ensembl transcriptome (release 109)63. Analysis of differential gene expression was performed using DESeq2 (v1.40.1). To identify differentially regulated genes between samples for downstream analyses, we selected genes with log2-transformed fold change greater than 0.7 or less than â0.7 and an adjusted P valueâ<â0.01. Gene ontology analysis was performed using topGO (v2.52.0) using the âweight01â algorithm for Fisherâs exact tests on R studio (version 2023.12.1â+â402). KEGG pathway analysis was performed using DAVID (https://david.ncifcrf.gov/tools.jsp)64,65.
Reanalysis of single-cell sequencing data
Previously published single-cell RNA sequencing data from gastrulating mouse embryos were accessed using MouseGastrulationData (v1.12.0) (https://github.com/MarioniLab/MouseGastrulationData). Data were subset to include only âepiblastâ, âprimitive streakâ and ânascent mesodermâ cell states from all samples staged E6.5 to E8.5 (samples 1â10, 12â 20 and 23â 37; samples 11, 21 and 22 were omitted from our analysis because their staging was ambiguous, for example, a sample labelled as âmixed gastrulationâ). Counts were log-normalized using Seurat (v. 4.3.0)66,67,68,69,70. Cells were first randomly down-sampled for visualization purposes to 400 cells per cell type. Pseudotime and trajectory interference analyses were performed using Slingshot (v2.8.0)71 for principle curve calculation; SingleCellExperiment (v1.22.0)72 and scater (v.1.28.0)73 were used for visualization of gene expression over pseudotime. The principle curve was then traced over the first two principal components to infer pseudo-temporal organization.
Statistics and reproducibility
Statistical tests were performed on GraphPad Prism 10.2.3 software. For comparison of two groups, two-sided unpaired t-tests were applied. For comparisons of three or more groups, one-way ANOVAs were used, followed by Dunnettâs multiple comparison tests for comparisons to control or Tukeyâs multiple comparison tests for comparisons between all groups, unless otherwise stated. P values are displayed on figure panels or in legends. All the experiments were performed at least in three biological replicates unless specifically described in the methods and the figure legends. Figure legends indicate the number of embryos, cells, explants or stem cell structures and, when relevant, the number of experiments performed for each analysis. Statistical power calculations were not used to determine sample size.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.