Mice
In all experiments, male C57BL/6J-OlaHsd mice between 8 and 12 weeks of age were used. Mice were bred in-house under specific-pathogen-free conditions in accordance with Irish and European Union regulations. Il17a/fâ/â mice were received from the laboratories of V. K. Kuchroo and Y. Iwakura. Il17aâ/â mice were received from the laboratory of K.H.G.M. IL-17A-GFP mice were purchased from The Jackson Laboratory. The Vav1Cre, Il7rCre, Per1Venus and Arntlfl mouse lines were bred and maintained at the Champalimaud Centre for the Unknown (CCU) animal facility. All mouse work was performed in compliance with the L.L. laboratory project licence, with ethical approval from the Trinity College Dublin ethics committee and the Animal Research Ethics Committee from the Health Products Regulatory Authority (HPRA), and the Brigham and Womenâs Hospital Institutional Animal Care and Use Committee guidelines. Experiments involving the Vav1Cre (ref. 55), Il7rCre, Per1Venus (ref. 56) and Arntlfl (ref. 57) mouse lines were approved by national and local institutional review boards, the Direção Geral de Veterinária and CCU ethical committees. For experiments using rIL-17A, mice were administered 100âµl of rIL-17A (2âµg per mouse) or PBS control by intraperitoneal (i.p.) injection three times over five days.
Environmental modulation
Inversion of lightâdark cycles
For experiments manipulating light cycles, mice were placed inside a ventilated, light-tight cabinet at room temperature, and light was adjusted such that control mice had lights on from 07:00â19:00, and test mice had lights on from 19.00â07:00.
Diet models
For HFD feeding experiments, mice were fed a 45% or 60% HFD for 3 weeks or 16 weeks ad libitum, compared with control mice that were fed a SFD, in which 10% of the calories were from fat. For reverse-feeding experiments, mice were fed a SFD ad libitum from 07:00â19:00 during the light cycle, compared with controls that were fed a SFD ad libitum from 19:00 to 07:00 during the dark cycle. For isocaloric feeding experiments, mice were placed into metabolic cages, and food consumption was measured over two days. The total calories per day was calculated, and mice were then fed 50% of the total calories consumed per day, for one week, compared with ad-libitum-fed controls that had 24-h access to food. For high-sucrose feeding experiments, mice were provided a 20% sucrose solution as their drinking water ad libitum for three weeks.
Tissue processing
Mice were euthanized by CO2 inhalation and adipose tissue was removed, minced with a razor and digested in 1âmgâmlâ1 collagenase type II (Worthington) dissolved in RPMI, in a shaking incubator for 25â30âmin at 37â°C. Digested cells were filtered through a 70-μm nylon mesh and centrifuged at 300g for 5âmin to pellet the stromovascular fraction (SVF). Lymph nodes were disrupted through a 70-μm filter and centrifuged for 5âmin at 300g to pellet. After processing, the adipose SVF and lymph-node leukocytes were stimulated with phorbol 12-myristate 13-acetate (PMA; 10ângâmlâ1; Sigma), ionomycin (500ângâmlâ1; Sigma) and brefeldin A (BFA; 5âµgâmlâ1; Sigma) in cRPMI medium, and incubated at 37â°C for 3âh. For cytokine stimulations, adipose SVF was stimulated with rIL-23 (10ângâmlâ1; Miltenyi) and rIL-1β (10ângâmlâ1; Immunotools) with BFA (5âµgâmlâ1) for 3âh.
For the circadian cytokine production of lymph-node γδ T cells, all lymph nodes were collected from mice at 07:00 (ZT0) and plated at 3-h time points over 24âh. Each time point was then stimulated with PMA and ionomycin in BFA for 3âh, leading up to the respective time point, before being collected for fluorescent staining for flow cytometry analysis.
For treatments with the REV-ERBβ agonist SR9009 (Sigma), cells were stimulated with PMAâionomycin or with the recombinant cytokines rIL-1β and rIL-23 as described above, in the presence or absence of SR9009. For doseâresponse graphs, lymph-node cells were stimulated in the presence of 0âµM, 5âµM, 10âµM, 20âµM, 40âµM and 80âµM doses of SR9009. Adipose SVF was stimulated with 40âµM of SR9009 only.
ELISA
γδ T cells were pre-expanded in wild-type mice using IL-7 and IL-15 cytokines and sorted to purity. A total of 50,000 γδ T cells were adoptively transferred into Tcrδâ/â mice. After one day, wild-type, Tcrδâ/â and Tcrδâ/â age-matched mice reconstituted with 50,000 γδ T cells were euthanized and adipose tissue lysates were assayed for IL-17A protein levels by ELISA. Processed adipose SVF lysates were diluted 1:2 in reagent diluent (1% bovine serum albumin in PBS) and IL-17A protein levels were quantified using the Mouse IL-17 Quantikine ELISA kit (M1700, R&D Systems).
Culturing of mouse adipocytes
Interscapular BAT was isolated, minced and incubated in Dulbeccoâs modified Eagleâs medium (DMEM) (glucose-free) supplemented with 10% fetal calf serum (FCS), 2.5 mM l-glutamine and 5âmM glucose. Tissue explants were then treated with or without rIL-17A (50ângâmlâ1; R&D Systems) for 18âh at 37â°C. Once the medium was removed, explants were snap-frozen in liquid nitrogen for RNA extraction.
Flow cytometry analysis
For intracellular and intranuclear staining, cells were washed with 1âml PBS and incubated in ZombieAqua Cell Viability Dye (Biolegend; 1:1,000 in PBS) for 20âmin at room temperature. Cells were incubated with an extracellular fluorochrome-labelled antibody cocktail with Fc block (1:200; BD Biosciences) in FACS buffer (PBSâ+â2% FCS) for 20âmin at room temperature. Cells were then washed with 2% FACS buffer resuspended in 100âµl of Foxp3 staining kit (eBioscience) and incubated at room temperature for 20âmin. Cells were washed with 1à permeabilization buffer (eBioscience) and then incubated with an intracellular fluorochrome-labelled antibody cocktail in 1à permeabilization buffer (eBioscience) for 20âmin at room temperature. Cells were incubated on ice for 30âmin and subsequently washed in 2% FACS buffer. Cells were acquired on a FACS Canto or LSR Fortessa cytometer (BD Biosciences). Data were analysed with FlowJo v.10 software. Cell sorting was performed using a FACSAria (BD Biosciences). Sorted populations were more than 95% pure. For a list of flow cytometry antibodies, see Supplementary Table 1.
Induction and assessment of EAE
Mice were fed either ad libitum or a 60% HFD three weeks before EAE induction. EAE was induced in male mice by subcutaneous immunization with MOG35â55 peptide (150âµg per mouse; GenScript) emulsified in complete Freundâs adjuvant (Condrex) containing 4âmgâmlâ1 heat-killed Myocobacterium tuberculosis (MTB). Mice were injected i.p. with pertussis toxin (200âng per mouse) (Kaketsuken) on day 0 to induce EAE development. Disease severity was monitored and assessed by clinical scores as follows: no clinical signs, 0; limp tail, 1; ataxic gait, 2; hind limb weakness, 3; hind limb paralysis, 4; tetra paralysis or moribund, 5. A weight loss of more than 20% constituted an additional humane end-point.
RNA extraction and quantitative PCR analysis
RNA extraction from isolated cells
Adipose γδ17 T cells were isolated and sorted from adipose tissue. RNA and cDNA were generated from isolated cells using the SYBR Green Fast Advanced Cells-to-CT Kit (Invitrogen) following the manufacturerâs specific instructions. To quantify the relative mRNA expression of genes of interest, quantitative PCR was performed in a 96- or 384-well plate format (Thermo Fisher Scientific) using PowerUp SYBR Green Master Mix (Invitrogen)-based detection (eBioscience). Relative mRNA levels were calculated using the ââ cycle threshold (ââCt) method and normalized to corresponding endogenous controls (Actb).
RNA extraction from tissues
Tissues were snap-frozen in liquid nitrogen, defrosted at room temperature and transferred to a 2-ml tube containing a 5-mm stainless steel bead. Tissues were homogenized in 1âml trizol reagent (Thermo Fisher Scientific) in a tissue lyser for 2.5âmin, 25 pulses per second. Then, 200âµl chloroform was added to each tube, and they were inverted once and left at room temperature for 2â3âmin, before centrifuging at 12,000g for 15âmin. The aqueous phase containing RNA was transferred into a new Eppendorf tube and 500âµl isopropanol was added to precipitate the RNA. Tubes were inverted ten times and left at room temperature for 10âmin, and then centrifuged at 12,000g for 10âmin. Supernatants were discarded and RNA pellets were washed in 1âml 75% ethanol, diluted in RNAse free dH2O. Tubes were centrifuged at 12,000g for 5âmin and supernatants were discarded by inverting the tube. The RNA pellet was left to dry at room temperature for 20â30âmin, until transparent, and the pellets were resuspended in 50âµl RNAse free water. RNA was left on ice for 30âmin, then in a heat block set at 55â°C for 15âmin. RNA quality and concentration were determined using a Nanodrop 2000 UV spectrophotometer (Thermo Fisher Scientific). Twenty microlitres of cDNA was synthesized from 2âµg of isolated RNA using the High-Capacity cDNA Reverse Transcription Kit (Biosciences) in a MiniAmp Thermal Cycler (BD Biosciences). To quantify the relative mRNA expression of genes of interest, quantitative PCR was performed in a 96- or 384-well plate format (Thermo Fisher Scientific) using SYBR Green-based detection (eBioscience). Relative mRNA levels were calculated using the ââ cycle threshold (ââCt) method and normalized to corresponding endogenous controls (Ppib or Rpl18). For a list of primers, see Supplementary Table 2.
Protein analysis by western blotting
BAT from wild-type and Il17aâ/â mice was collected over 24âh and snap-frozen. The BAT was then lysed and centrifuged at 14,000g, and the pellet was discarded. The amount of protein was quantified using a BCA kit. Tissue lysates were subsequently boiled at 95â°C for 5âmin to denature the proteins. Proteins were resolved, on the basis of their molecular weight, through SDSâPAGE gels in 1à running buffer. Proteins were electro-transferred onto PVDF membranes (Merck) in 1à transfer buffer containing 10% methanol. Membranes were blocked in 5% milk in 1à Tris-buffered saline with Tween-20 and incubated with SCD1, or α-tubulin primary antibodies (Cell Signaling Technologies) overnight at 4â°C and HRP-conjugated secondary antibodies (Jackson Immunoresearch). Membranes were incubated in ECL substrate (BioRad), and images were developed on a Biorad Gel Doc imaging system. Images were quantified using ImageJ. For raw uncropped blots, see Supplementary Fig. 1.
Plasma 2H2O enrichment analysis and DNL calculations
For measurements of DNL in vivo, wild-type or Il17a/fâ/â mice were placed overnight on 8% enriched D2O drinking water, with subsequent collection and snap-freezing of BAT and liver. The 2H labelling of water from samples or standards was determined by deuterium acetone exchange, and using equations as previously described58.
Metabolic cage analysis
Indirect calorimetry experiments were performed with the Promethion metabolic cage system, or the comprehensive lab animal monitoring system (CLAMS), essentially as described59. Mice were singly housed and allowed at least 12âh of acclimatization to the new environment. O2 consumption, CO2 emission, energy expenditure, body weight, food and water intake and locomotor activity were monitored throughout the experiment. For experiments involving the manipulation of light cycles, mice were placed inside the metabolic cages at room temperature, and light was adjusted such that the control mice had lights on from 07:00â19:00, and test mice had lights on from 19:00â07:00. Analysis was performed using the online indirect calorimetry vignettes CalR (ref. 60); online software is available at https://calrapp.org/.
Bulk RNA-seq and scRNA-seq data analysis
The bulk RNA-seq dataset of BAT from wild-type and AdIl17rcâ/â mice was downloaded from the GEO repository GSE144255 as cuff gene counts22. The bulk RNA-seq datasets of skin biopsies from patients with psoriasis who were receiving brodalumab36 and ixekizumab37 were downloaded from the GEO repositories GSE117468 and GSE31652, respectively. Heat maps were generated using the online platform Heatmapper61 (http://heatmapper.ca/).
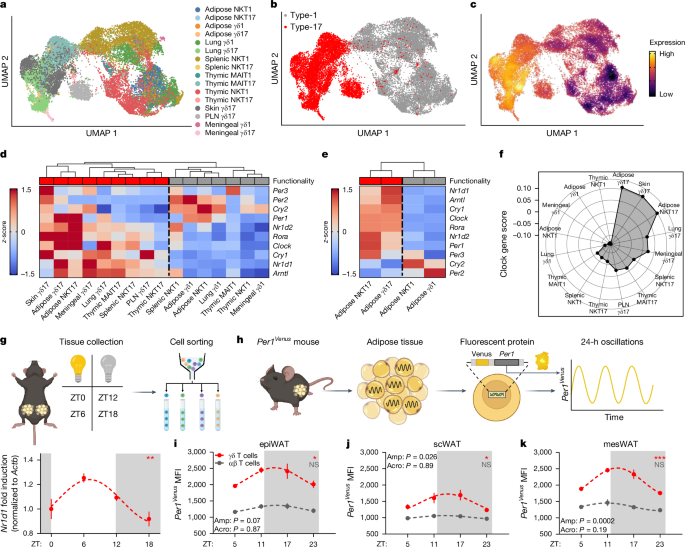
scRNA-seq was performed on single-cell suspensions of sorted γδ T cells and iNKT cells from the visceral adipose tissue of C57BL/6J mice using the 10x Genomics platform. A total of five visceral adipose tissue deposits from five independent biological replicates were pooled for sequencing. Single-cell suspensions were loaded onto a 10x Chromium Controller to generate gel beads-in-emulsion (GEMS), and GEMs were processed to generate unique molecular identifier (UMI)-based libraries according to the 10X Genomics Chromium Single Cell 3â protocol. Libraries were sequenced using a NextSeq 500 sequencer. Raw BCL files were demultiplexed using Cell Ranger v.3.0.2 mkfastq to generate fastq files with default parameter. Fastq files were aligned to the mm10 genome (v.1.2.0) and feature reads were quantified simultaneously using the Cell Ranger count for feature barcoding. Filtered feature-barcode UMI count matrices containing quantification of gene expression were used for downstream analysis.
Downstream scRNA-seq data analysis
A total of 22,748 cells mouse γδ T cells, iNKT cells and MAIT cells expressing a median of 1,423 genes and 3,556 UMIs per cell were loaded from feature-barcode UMI count matrices using the Seurat v.4.1.0 package62. Adipose and splenic iNKT cells were reanalysed from GSE142845 (ref. 63), thymic iNKT cells were reanalysed from GSE141895 (ref. 64), thymic MAIT cells were reanalysed from E-MTAB-7704 (ref. 65), pulmonary γδ T cells were reanalysed from E-MTAB-8732 (ref. 66), peripheral lymph node (PLN) and dermal γδ T cells were reanalysed from GSE123400 (ref. 67) and meningeal γδ T cells were reanalysed from GSE147262 (ref. 68). Cells expressing more than 11% of mitochondrial genes as a percentage of total gene counts were considered to represent apoptotic or dead cells and were therefore removed from the analysis. Cells were also filtered on the basis of total UMI counts and total gene counts on a per-sample basis to remove empty droplets, poor quality cells and doublets, with a minimum cut-off of at least 300 genes per cell across all samples. UMI counts were normalized using regularized negative binomial regression using sctransform v.0.3.3 (ref. 69).
Dimensionality reduction was performed using principal component analysis (PCA) with nâ=â100 dimensions and 2,000 or 3,000 variable features, and an elbow plot was used to determine the number of PCA dimensions used as input for UMAP70. For collective analysis of cells from different batches and cells sequenced using different scRNA-seq technologies the Harmony v.1.0 package71 was used with default settings to remove batch effects, and batch-corrected harmony embeddings were used for UMAP. UMAP was performed using a minimum distance of 0.3 and a spreading factor of 1. Shared nearest neighbour graphs were calculated using kâ=â20 nearest neighbours. Graph-based clustering was performed using the Louvain algorithm. In some cases, overclustering was performed and clusters were manually collapsed, and/or the first two dimensions of the UMAP reduction were used as input for graph-based clustering instead of PCA or harmony embeddings. Adipose γδ T cells were distinguished from adipose iNKT cells by expression of Trdc and graph-based clustering, and analysed separately for γδ1 versus γδ17 cell identification. Type-1 and type-17 innate T cells were individually identified in each dataset using graph-based clustering and gene-expression analysis, and cycling cells were excluded. For thymic innate T cells, CD44â progenitor populations were also excluded. Raw counts from type-1 and type-17 cell populations were then merged and normalized together into a single file for comparative analysis.
Gene-expression analysis was performed using the FindMarkers() or FindAllMarkers() Seurat functions and the Wilcoxon rank sum test. Heat maps were generated using the Complex Heatmap v.2.7.11 and circlize v.0.4.14 packages72, and module scores were calculated using the AddModuleScore() Seurat function with nâ=â10 control features. Density plots were produced using the Nebulosa v.1.1.1 package73. Log-normalized RNA counts were used for all gene-expression analysis and visualization. All computational analysis was performed using R v.4.1.2 and RStudio Desktop v.1.4.1712 on an Ubuntu 20.04 Linux GNU (64âbit) system.
Statistical analysis
GraphPad Prism 9.3.0 was used for statistical analysis. For all experiments, a 95% confidence interval was used and Pââ¤â0.05 was considered statistically significant. A DâAgostinoâPearson omnibus normality test was first performed to test whether the data were normally distributed (Gaussian distribution). If data were normally distributed, parametric testing was performed. If data were not normally distributed, non-parametric testing was performed. When comparing two groups, an unpaired or paired two-tailed studentâs t-test was used. When comparing more than two groups, an ordinary one-way ANOVA with Dunnettâs test was used. When comparing data with two variables, a two-way ANOVA with Bonferroni test was used. Cosine curves were fitted in GraphPad Prism and circadian rhythmicity was evaluated using the cosinor regression model using the cosinor and cosinor2 R packages in RStudio. The circadian period was assumed to be 24âh for all analysis and the significance of the circadian fit was assessed by a zero-amplitude test with 95% confidence. Amplitude and acrophase were extracted from the cosinor model. In all figures, *Pââ¤â0.05, **Pââ¤â0.01, ***Pââ¤â0.001, ****Pââ¤â0.0001.
Graphical representations
All diagrams and graphical representations were created using BioRender.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.