Mice
Miclâ/â and C57BL/6J mice were bred and maintained under specific pathogen-free (SPF) conditions at the University of Aberdeen, University of Exeter and Charles River Laboratories. Mice were housed in separate groups with bedding exchanges between cages every 2 days for 1 week before commencement of experiments, and maintained on a 12âhâ12âh darkâlight cycle (07:00â19:00) at 20â24â°C and relative humidity of 55â±â15%. Mouse experiments were performed by random assignation of age-matched (6â8 weeks old) and sex-matched mice in experimental or control groups at the beginning of each experiment; females were co-housed and experiments were not blinded. All experiments conformed to the ethical review committee of the University of Aberdeen, University of Exeter and the UK Home Office regulations (project license numbers: P79B6F297 and P6A6F95B5).
CAIA
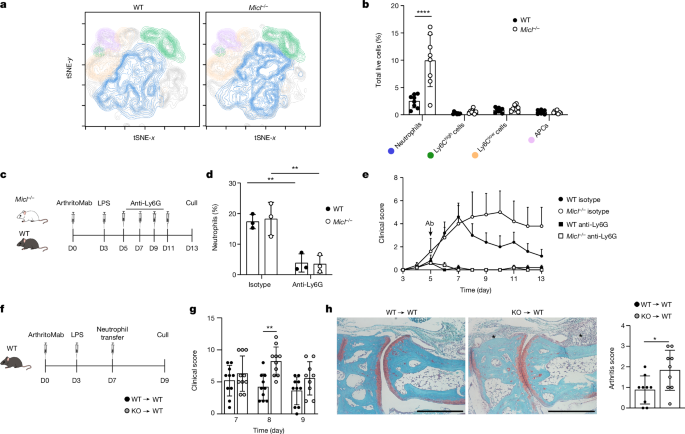
Male mice received intraperitoneal (i.p.) injections of 2âmg of ArthritoMab monoclonal antibody cocktail (MD Bioproducts) on day 0, followed by 50âμg of lipopolysaccharide (LPS) i.p. (MD Biosciences) on day 3. Joint inflammation was scored visually using a scale of 0 (no visible signs of redness or swelling) to 4 (extensive swelling with signs of deformity). To achieve neutrophil depletion during CAIA, 500âμg of rat anti-mouse Ly6G (clone 1A8, Bio-X-Cell) or isotype control (rat anti-mouse IgG2a; Bio-X-Cell) was administered i.p. to mice every 48âh from day 5 onwards. Alternatively, 50âμg of rat anti-mouse GR-1 (clone RB6-8C5) or isotype control (rat anti-mouse IgG2b) was administered i.p. every 48âh from day 5 onwards. Mice were culled on day 5, 7, 10, 11 or 13, as indicated in the text. For adoptive transfer experiments, bone marrow neutrophils were isolated as detailed below and stained with cell proliferation dye eFluor 670 (eBioscience). Labelled cells (5âÃâ106) were transferred intravenously (i.v.) to WT mice on day 7 following induction of CAIA. Mice were culled on day 9. To investigate the role of NET formation, mice undergoing CAIA were injected i.p. with 2âmgâkgâ1 daily of BB-CL-amidine (Cayman Chemical), 4âmgâkgâ1 daily of GSK484 (Cambridge Biosciences), 75âU per animal of DNase I (Merck) or vehicle (5% DMSO in 10% cyclodextran for BB-Cl-amidine or ethanol 99.9% diluted 1:50 in 0.9% NaCl for GSK484) from day 7 to day 10. Mice were culled on day 11.
K/BxN serum transfer model of arthritis
Male mice were administered i.p. with 100âμl of serum from transgenic K/BxN mice26. The development of clinical symptoms were monitored daily and mice were culled on day 10. Joint inflammation was scored visually using a scale of 0 (no visible signs of redness or swelling) to 4 (extensive swelling with signs of deformity). To investigate the role of NET formation, mice were injected i.p. with 4âmgâkgâ1 daily of GSK484 or vehicle (ethanol 99.9% diluted 1:50 in 0.9 % NaCl) from day 7 to day 10. Mice were culled on day 11.
CIA
Male DBA/1 mice were purchased from Inotiv and maintained at the University of Exeter. Mice were treated subcutaneously with 100âμl of Immunization Grade Chick Type II Collagen (Chondrex; 200âµg per mouse) in Complete Freundâs Adjuvant (Sigma-Aldrich) at day 0 and with 100âμl of Immunization Grade Chick Type II Collagen (200âµg per mouse) in Incomplete Freundâs Adjuvant (Sigma-Aldrich) at day 14, followed by an i.p. injection of 50âµg of LPS at day 26. Of anti-MICL antibodies, 0.7âmg of isotype control antibodies or PBS were administered i.p. every 48âh from day 17 to day 33. Mice were culled at day 35. In our animal facility, naive DBA/1 animals did not develop any spontaneous form of arthritis by 13 weeks of age (the latest timepoint in our experiments).
Histology
For histology, paws were fixed in 4% paraformaldehyde (PFA) overnight at 4â°C and decalcified in 10% EDTA for 3â4 weeks. Decalcified paws were embedded in optimal cutting temperature (OCT) and cryosectioned or embedded in paraffin wax and sectioned before staining with haematoxylin and eosin, or Safranin O, haematoxylin and fast green. Scoring of histological sections was performed blinded using an arthritis severity score as previously described18.
Immunofluorescence staining protocol
Frozen tissue sections were thawed for 30âmin at room temperature and washed with PBS before permeabilization with 0.25% Triton X-100 or 0.5% saponin in PBS for 10âmin. After permeabilization, sections were washed with PBS again and blocked with 3% BSA at room temperature for 30âmin. Tissue sections were stained with primary antibodies (anti-cit-H3 (Abcam), anti-DNA/H1 (Merck) and anti-GR-1 (produced in-house59)) diluted in blocking buffer (3% BSA in PBS) for 1âh 30âmin in a humidified chamber, after which they were washed with PBS-Tween-20 (0.05%) and stained with secondary antibodies for 1âh at room temperature in the dark. Sections were washed as before and counterstained with 1âμgâmlâ1 DAPI and mounted in Vectashield antifade mounting medium for fluorescence (Vectorlabs). Coverslips were sealed with nail polish and slides were stored in the dark at 4â°C until imaging. Fluorescence was visualised using the Zeiss confocal LSM700 microscope and Zen Black software (Zeiss). The quantification of positive area (area+ %) for each channel was conducted using Fiji software. Image preprocessing included utilizing the built-in âMomentsâ algorithm for thresholding each channel.
Neutrophil isolation
Bone marrow neutrophils were isolated using Histopaque (Merck) by a density gradient centrifugation method or using the EasySep Mouse Neutrophil Enrichment Kit (StemCell Technologies) according to the manufacturerâs guidelines. Flow cytometry analysis confirmed an approximately 90% pure neutrophil population (the remaining population consisting primarily of monocytes). To isolate thioglycollate-elicited neutrophils, mice were injected i.p. with 1âml of 3% thioglycollate broth. After 4âh, mice were culled and neutrophils were harvested by peritoneal lavage with PBS 5âmM EDTA.
Human neutrophils from the blood of healthy donors were purified using a Ficoll-Paque density centrifugation method60 or using the EasySep direct human neutrophil isolation kit (StemCell Technologies) as per the manufacturerâs instructions. Samples were obtained from consenting healthy donors with the approval of the Faculty of Health and Life Sciences ethics review board, University of Exeter (eCLESBio000371) and the College of Life Sciences and Medicine ethics review board, University of Aberdeen (CERB number 1243).
CD34+ cell isolation and CRISPR-mediated knockout
Peripheral blood mononuclear cells (PBMCs) were isolated from apheresis blood waste (NHSBT) with ethical approval from NHS Research Ethics Committee (REC 18/EE/0265) by density centrifugation using Histopaque-1077 (Sigma-Aldrich) according to the manufacturerâs instructions. After washing, cells were resuspended in red cell lysis buffer (55âmM NH4Cl, 0.137âmM EDTA and 1âmM KHCO3, pH 7.5) and incubated on ice for 10âmin. Next, to enrich haematopoietic stem cells, CD34+ cells were isolated from PMBCs using a human CD34 Microbead Kit (Miltenyi Biotec) according to the manufacturerâs protocol. Isolated cells were cultured in Iscoveâs modified Dulbeccoâs medium (IMDM) supplemented with 10% (v/v) FBS and 1% (v/v) penicillinâstreptomycin at 37â°C in 5% CO2. Cytokines were added at the indicated concentrations and days of culture: stem cell factor (SCF; 50ângâmlâ1; day 0â5 of culture), Flt-3 ligand (50ângâmlâ1; day 0â5 of culture), interleukin-3 (IL-3; 10ângâmlâ1; day 0â5 of culture), granulocyteâmacrophage colony-stimulating factor (GM-CSF; 10ângâmlâ1; day 3â7 of culture) and granulocyte colony-stimulating factor (G-CSF; 10ângâmâ1; day 7â14 of culture). All functional assays were completed between day 17 and day 18 of culture49.
CRISPR-mediated knockout was completed using the AmaxaTM 4D-Nucleofector (Lonza) using a P3 Primary Cell 4D-NucleofectorTM X Kit S (Lonza) and TrueCut Cas9 Protein v2 (Invitrogen) according to the manufacturersâ instructions. In brief, day 3 cultured neutrophils were resuspended in the mixture of P3 Primary Cell Solution and supplement 1 containing 50âpmol Cas9 and 125âpmol of guide RNA (62.5âpmol of two guides with the same gene target). Cells were transferred into nucleocuvette strip and electroporated using the EO-100 program. After electroporation cells were transferred into six-well plates containing StemSpan medium (StemCell Technologies) supplemented with FBS and penicillinâstreptomycin and containing SCF, Flt-3 and IL-3. From day 5 onwards, cells were cultured as outlined above in IMDM. All functional assays were completed between day 17 and day 18 of culture. Guide RNAs were designed using Knockout Guide Design (Synthego). The following single guide RNAs were used (Synthego, modified single guide RNA with EZ scaffold): CLEC12Aâ+â9979452: UGAAUAUCUCCAACAAGAUC and CLEC12A-9979406: GUUGUAGAGAAAUAUUUCUC; negative control scrambled #1: GCACUACCAGAGCUAACUCA and negative control scrambled #2: GUACGUCGGUAUAACUCCUC. Cells were stained with anti-CD66-PE/Dazzle (clone G10F5; diluted 1:50), anti-CD15-AF700 (clone Hi98; diluted 1:50), anti-CD16-APC (clone 3G8; diluted 1:50) and anti-hMICL or isotype control antibodies at 10âµgâmlâ1 to confirm loss of MICL expression.
ROS
Bone marrow neutrophils and human neutrophils were isolated as described above and resuspended in OptiMEM (Thermo Fisher Scientific) supplemented with 5% FCS. Cells (5âÃâ105) were added to each well of a white 96-well flat-bottomed plate and stimulated in triplicate with 200âμgâmlâ1 MSU crystals (Invivogen), 25âμgâmlâ1 Zymosan (Molecular Probes), 100ânM PMA (Sigma), isolated NETs (100âμgâmlâ1 based on NETâDNA concentration) or A. fumigatus hyphae (1âÃâ104 conidia per well incubated for 12âh at 37â°C) in the presence of 100âμM luminol (Sigma). Chemiluminescence was measured every 3âmin for 2âh in a FLUOStar Optima microplate reader (BMG Labtech) or a Spark Cyto (Tecan) at 37â°C with 5 % CO2.
NETs
Bone marrow and thioglycollate-elicited neutrophils were isolated as described above. Cells were resuspended in RPMI (without phenol red; Thermo Fisher Scientific) supplemented with 2% DNaseâ/â mouse serum and seeded in an eight-well iBidi μ-slide (iBidi). Cells were stimulated with 100âμgâmlâ1 of MSU crystals (Invivogen), 100ânM PMA (Sigma), isolated NETs (100âμgâmlâ1 based on NETâDNA concentration) or A. fumigatus hyphae and incubated at 37â°C with 5% CO2 for 4âh. In some experiments, as indicated in the text, DPI (10âμM; Sigma) or NSC-87877 (5âμM; Cayman Chemical) were included in the assays. Extracellular DNA was visualized with 5âμM Sytox Green (Invitrogen). In experiments without fixation, cells were previously stained with Draq5 (Thermo Fisher Scientific) and cell-impermeable Sytox Green, and images were acquired on an inverted Zeiss AxioObserver Z1 using a PlanApo Ã20/0.75 NA dry lens (Carl Zeiss) and a Hamamatsu Fusion sCMOS camera with an attached incubation chamber (PeCon GmbH) at 37â°C. Fluorescent images were analysed in Image J, and NETs defined as Sytox Green-positive cells showing extrusions were counted.
NET (%)â=â(total Sytox Green-positive cells extruding NETs/total cells counted)âÃâ100
For kinetic curves of NET formation, neutrophils were seeded in a 96-well black plate with a transparent bottom and the cells were left to adhere for 30âmin in a cell culture incubator. Cells were stimulated with the defined reagents and 5âμM Sytox Green. Fluorescence signal (504-nm excitation and 523-nm emission) was measured every 10âmin for 7âh in a Spark Cyto (Tecan) at 37â°C with 5% CO2.
NETs were isolated as previously described61. In brief, bone marrow-isolated or purified human neutrophils were plated in a six-well plate at a density of 1âÃâ106 cells per well in RPMI without phenol red. Following stimulation with 100ânM PMA for 4âh at 37â°C, the culture medium was removed and NETs were partially digested by application of a restriction enzyme mix combining the enzymes BseRI, PacI, NdeI and AfIII (New England Biolabs) at a concentration of 5âUâmlâ1 in NEB buffer for 1âh at 37â°C. Supernatants were collected and centrifuged at 300g for 10âmin at 4â°C. NET supernatants were transferred to a fresh tube and stored at â80â°C until used. When indicated, NETs were treated with DNase I (Thermo Fisher Scientific) or proteinase K (Roche) for 1âh at 37â°C.
Recognition of isolated NETs by MICL was assessed by ELISA. A 96-well plate (Nunc Maxisorp) was coated with NETs diluted in PBS overnight at 4â°C. Wells were washed and blocked with blocking buffer (5% BSA) at room temperature. FcâmMICL or FcâmCLEC12B at 1âμgâmlâ1 in blocking buffer was added and incubated for 2âh at room temperature. Bound Fc-fusion proteins were detected with horseradish peroxidase-conjugated goat anti-human IgG (Jackson Immunoresearch) diluted 1:10,000 for 1âh. TMB substrate was added, and absorbance was measured using a plate reader (Tecan). Purification of FcâMICL and FcâmCLEC12B, from transduced HEK293T cells (originally purchased from the American Type Culture Collection (ATCC), but not tested for Mycoplasma contamination for the experiments detailed in this paper), was performed as previously described32.
To analyse the recognition of isolated NETs by MICL using a MICL-expressing cell line, a 96-well plate was coated with NETs diluted in PBS overnight at 4â°C. Wells were washed, and BWZ.36 NFAT-LacZ cells (provided by W. Yokoyama; not tested for Mycoplasma contamination for the experiments detailed in this paper) expressing the CD3ζ chain fused to the transmembrane and carbohydrate recognition domain (CRD) of mMICL or mCLEC12B were added (2âÃâ105 cells per well) for 18âh at 37â°C. After stimulation, cells were centrifuged at 800g for 2âmin and washed two times with PBS. Of CPRG substrate buffer, 100âµl was added per well. The reaction was stopped 4âh later with the addition of 50âµl of glycine-EDTA buffer, and the absorbance was measured using a plate reader62 (Tecan). Antibody crosslinking with the appropriate receptor antibody was used to confirm the functionality of the chimeric receptor constructs.
To analyse neutrophil infiltration by isolated NETs, 6â8-week-old Miclâ/â and C57BL/6J mice were injected i.p. with 300âμg NETs or 50âμg LPS. Four hours later, mice were euthanized, and their peritoneal cells were counted and analysed by flow cytometry. Cell-free DNA was evaluated using the Quant-iT PicoGreen dsDNA Assay (Invitrogen) following the manufacturerâs instructions.
Immunofluorescent staining of in vitro samples
Neutrophils (5âÃâ104) were plated in an eight-well iBidi μ-slide (iBidi) 1âh before stimulation with isolated NETs, PMA or MSU. Four hours after stimulation, cells were fixed for 20âmin at 37â°C with 2% PFA and permeabilized with 0.5% Triton X-100 in PBS. Samples were blocked with 3% (v/v) normal goat serum, 1% (w/v) BSA in PBS and incubated with anti-cit-H3 (Abcam) and anti-myeloperoxidase (R&D) antibodies. The secondary antibodies donkey anti-rabbit Cy5 and rabbit anti-goat Alexa Fluor 488 were used. Finally, the samples were stained with DAPI.
Fluorescence quantification was conducted by segmenting the DAPI channel using QuPath software (version 0.4.3) with the Cellpose extension (https://github.com/BIOP/qupath-extension-cellpose). Before segmentation, preprocessing of the DAPI channel was carried out using Fiji software to mitigate background noise, involving background subtraction and enhanced contrast built-in functions.
Preprocessing of the GFP and Texas Red channels involved a background subtraction of the mean grey value and two times the standard deviation of an empty region (background noise). The quantification of each channel was conducted using the measurement function of Fiji software, utilizing the mask generated by Qupath.
Flow cytometry and monoclonal antibodies
Cells were isolated from the hind paw ankle joint of arthritic mice using the protocol previously described20. Peripheral blood was collected by cardiac puncture or tail nicking in the presence of EDTA and red blood cell lysis performed using PharmLyse (BD Biosciences). Single-cell suspensions were stained with fixable viability dye eFluor 780 (eBioscience) and further stained with conjugated antibodies for same-day acquisition or fixed in 2% PFA. Conjugated antibodies used in these experiments included: anti-CD45-FITC (clone 102), anti-CD45-PerCP-cyanine5.5 (clone 102), anti-CD11b-BUV395 (clone M1/70), anti-CD11b-PE-Cy7 (clone M1/70), anti-GR-1-APC (clone RB6-8C5), anti-MHC-II-FITC (clone 2G9), anti-MHC-II-BUV496 (clone 2G9), anti-C5aR-BV510 (clone 20/70), anti-Ly6G-BV421 (clone 1A8), anti-Ly6G-Spark Blue 550 (clone 1A8), anti-Ly6G-APC (clone 1A8), anti-CD62L-BV510 (clone MEL-14), anti-CD18-BV650 (clone C71/16), anti-CD18-APC (H155-78), anti-F4/80-AF700 (clone BM8), anti-F4/80-PE-Cy7 (clone BM8), anti-Ly6C-PE-Cy7 (clone HK1.4), anti-Ly6C-Brilliant Violet 570 (clone HK1.4), anti-CCR1-PE (clone 643854), anti-CXCR2-APC (clone SA045E1), anti-CD11c-BV711 (clone HL3), anti-B220-BV605 (clone RA3-6B2) and anti-CD3-Alexa Fluor 647 (clone 17A2). All were purchased commercially from eBioscience, R&D systems or BioLegend and used diluted 1:600. Anti-mCLEC12A-biotinylated21, anti-hCLEC12A63 and isotype control AFRC MAC 49 (ECACC 85060404; isotype for anti-MICL) were generated in-house and used at 10âμgâmlâ1. Anti-mCLEC12A-biotinylated21 and anti-hCLEC12A63 antibodies were validated using mouse and human CLEC12A-transduced NIH3T3 fibroblast (originally purchased from the ATCC; not tested for Mycoplasma contamination for the experiments detailed in this paper). Cells were acquired using a BD LSR II Fortessa flow cytometer (BD Biosciences), BD LSRFortessa X-20 flow cytometer (BD Biosciences) or Cytek Aurora Spectral cytometer (Cytek). See Extended Data Fig. 1b for the cellular gating strategy. Data were collected using BD FACSDiva v8.0.3 (BD Biosciences) or SpectroFlo software v3.2.1 (Cytek) and analysed using FlowJo v10 software (BD Biosciences).
Imaging flow cytometry
Single cells isolated from the arthritic joint, as detailed above, were fixed in 1% PFA for 30âmin at 37â°C. Cells were washed with PBS 2âmM EDTA for 5âmin at 500g and stained with conjugated antibodies (detailed above), anti-DNA/histone 1 (Merck Millipore; 1.4âμgâmlâ1) and anti-cit-H3 (Abcam; diluted 1:300) for 1âh at room temperature. The secondary antibodies goat anti-rabbit APC (Molecular probes) and goat anti-mouse AF488 (Invitrogen) at 1âμgâmlâ1 were added for 1âh at 4â°C. Cells were washed and resuspended in 50âμl of PBS 2âmM EDTA before acquisition using the Amnis ImageStreamX MKII (Luminex) INSPIRE acquisition software. Files were analysed using IDEAS software v6.2 (Luminex).
Single cells were selected by plotting the area feature of brightfield channel 1 (BF1) versus the aspect ratio parameter of the same brightfield channel, which is the ratio of minor axis to the major axis of the applied mask, and describes the shape of the mask applied to the cells (Extended Data Fig. 7f). Focused cells were then selected by plotting the âGradient RMSâ feature of BF1 against the BF1 contrast parameter. Cells with high-gradient and high-contrast value were more in focus and chosen for further analysis. All focused cells were used in the analysis for the presence of the cellular fluorescence parameters. Fluorescence parameters were measured in channel 2 (DNA/histone H1-AF488), channel 11 (cit-H3-APC) and channel 7 (Ly6G-BV421) with magnification set to Ã40.
Serum collection from patients with rheumatoid arthritis
Serum samples from patients with rheumatoid arthritis were obtained from the SERA cohort42 (Extended Data Table 1). Sera from healthy controls were obtained by collection of whole blood in a BD vacutainer serum collection tube (BD Biosciences). The blood was allowed to clot for 15â30âmin at room temperature, after which the tubes were centrifuged at 2,000g for 10âmin in a 4â°C. Sera were removed, aliquoted and stored at â80â°C. Serum samples were obtained from consenting healthy donors with the approval of the College of Life Sciences and Medicine ethics review board, University of Aberdeen (CERB number 1243).
Serum collection from patients with COVID-19
Blood from 121 patients diagnosed with SARS-CoV-2 infection at Weill-Cornell Medicine between March and July 2020. Research on patients with COVID-19 was reviewed and approved by the Institutional Review Board of Weill-Cornell Medicine (New York Presbyterian and Lower Manhattan hospitals; #IRB 20-03021645 and #IRB 20-03021671). Informed consents were obtained from all enrolled patients and health-care workers by trained staff, and records were maintained in our research database for the duration of the study. All patients were classified as mild/moderate (nâ=â25) and severe (nâ=â66) disease according to oxygen requirements with mild/moderate disease defined as SARS-CoV-2 infection and less than 6âl of non-invasive supplementary oxygen to maintain SpO2â>â92%, and severe disease was defined as SARS-CoV-2 infection requiring hospitalization and received 6âl or more supplementary oxygen or mechanical ventilation. For controls, we used blood samples from 36 SARS-CoV-2-negative individuals collected by the JRI IBD Live Cell Bank Consortium at Weill-Cornell Medicine. Heparinized plasma and serum samples were aliquoted, heat-inactivated at 56â°C for 1âh and then stored at â80â°C.
Serum collection from patients with SLE
All patients with SLE in the study met the revised American College of Rheumatology criteria64 and the SLICC criteria65. Some patients had a history of biopsy-proven nephritis according to the International Society of Nephrology/Renal Pathology Society classification. Healthy female volunteers (with no family history of autoimmune disease) served as age-matched and ethnicity-matched controls. All patients provided informed consent, and samples used in this research project were obtained from the Imperial College Healthcare Tissue Bank (ICHTB). The ICHTB is supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London. The ICHTB is approved by Wales REC3 to release human material for research (22/WA/0214), and the samples for this project (ref: R13010a) were issued from sub-collection reference number IMM_MB_13_001.
ELISAs from human patients
Sera from patients with rheumatoid arthritis, COVID-19 and SLE and healthy controls were screened for the presence of MICL autoantibodies using a modified ELISA method25. In brief, Nunc Maxisorp 96-well plates were coated with equivalent amounts of the Fc-fusion proteins FcâhMICL and FcâmCLEC12B diluted in PBS overnight at 4â°C. Plates were blocked with 10% BSA and 10% HI goat serum (Merck) in PBS for 1âh. Sera from patients with rheumatoid arthritis, COVID-19 and SLE and healthy control were diluted to a 1:256 dilution in PBS and added to the pre-blocked plate and incubated for 2âh. Bound autoantibodies were detected with horseradish peroxidase-conjugated goat anti-human F(abâ²)2 fragment (Jackson Immunoresearch) diluted 1:50,000 in PBS for 1âh. TMB substrate was added, and absorbance was measured at 450ânm in a plate reader (Tecan).
A. fumigatus systemic infection model
Miclâ/â and C57BL/6J female mice were injected intravenously with 106 A. fumigatus ATCC 13073 conidia, as previously described66. Mice were culled when they lost 20% body weight or had become moribund. To investigate the role of NET formation, mice were injected i.p. with 4âmgâkgâ1 daily of GSK484 (Cambridge Biosciences) from day 1 to day 5. Organs were homogenized in PBS and used for the determination of fungal burdens and levels of inflammatory cytokines. Fungal burdens were determined by serial dilution onto potato dextrose agar plates and normalized to organ weights. Cytokines were measured by ELISA (BD Biosciences), as described by the manufacturer, and normalized to protein concentration.
Statistical analysis
Data are represented as meanâ±âs.d., unless otherwise indicated. All statistical analyses were performed using GraphPad Prism (v9, GraphPad Software) and depicted in the respective figure legends. For all experiments with two groups, two-tailed unpaired Studentâs t-tests (equal variances) or two-tailed MannâWhitney tests were used. One-way or two-way ANOVA (with equal variances) with correction for multiple comparisons was performed for experiments with more than two groups. All Pâ<â0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.