Patient consent
Patients profiled in this study are participants in the Genomic Answers for Kids (GA4K) program at the Children’s Mercy Research Institute (https://www.childrensmercy.org/childrens-mercy-research-institute/studies-and-trials/genomic-answers-for-kids/). Potential GA4K participants are informed of the program by clinicians in our health system. Participants met with a member of the study team to ensure any questions they had were answered as a part of the informed written consent into GA4K. Participants did not receive any form of compensation. Genome sequencing data collected through GA4K has been de-identified and deposited in dbGaP (accession: phs002206.v2.p1). Biospecimens collected through GA4K cannot be distributed as general research reagents, but can be shared in instances directly related to achieving diagnoses. The Institutional Review Board of Children’s Mercy Research Institute gave ethical approval for this work (studies #00002465 and #11120514). All methods were carried out in accordance with relevant guidelines and regulations.
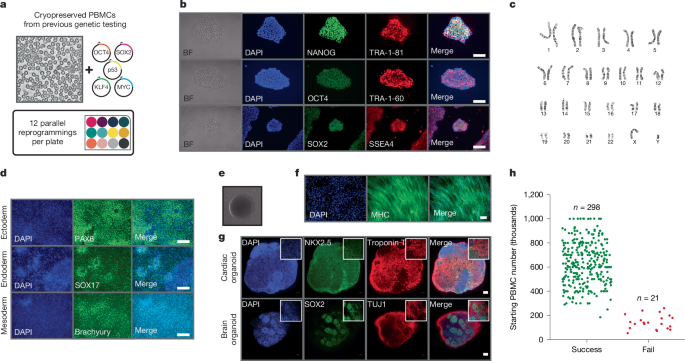
Generation of patient-derived iPS cell lines
Control human iPS cell line (WTC11 background) was obtained from Coriell Institute (GM25256; Supplementary Table 2). Patient iPS cells were generated from PBMCs. Cryopreserved PBMCs were thawed and cultured in StemSpan SFEM II (9605, StemCell Tech) supplemented with StemSpan erythroid expansion supplement (2692, StemCell Tech) and 1X antibiotic–antimycotic (15240062, Gibco) for 2–5 days before reprogramming using episomal plasmids. Episomal plasmids expressing OCT3/4, SOX2, KLF4, L-MYC, LIN28, p53 and EBNA1 (pCE-mP53DD (41856), pCXB-EBNA1 (41857), pCE-HUL (41855), pCE-hsk (41814), pCE-hOCT3/4 (41813; all Addgene) were introduced into PBMCs by electroporation using the Lonza 4D-Nucleofecter. The nucleofected PBMCs were transferred to a Matrigel (354277, Corning)-coated plate and maintained in StemSpan SFEM II plus 10 μM ROCK inhibitor. Two days after nucleofection stem cell media 1 (SCM1) composed of ReproTeSR (5926, StemCell Tech), 10 μM ROCK inhibitor (Y-27632 dihydrochloride; 1254, Tocris), 0.5 μM PD0325901 (72184, StemCell Tech), 6 μM CHIR99021 (72054, StemCell Tech), 2 μM SB431542 (72234, StemCell Tech), 250 μM sodium butyrate (72242, StemCell Tech), 50 μg ml−1 ascorbic acid (A8960, Sigma), 10 ng ml−1 bFGF (78003, StemCell Tech) and 1X antibiotic–antimycotic were added to each well without removing any media, centrifuged at 50g for 30 min and placed back in the incubator for 2 days. After 2 days, media were removed and fresh SCM1 was added. The next day, SCM1 was added without removing media; the following day, all media were removed and fresh SCM1 was added. This was repeated for 1 week. After week 1, media were changed to SCM2 composed of SCM1 minus SB431542, and media were changed exactly as described for SCM1. iPS cell colonies appeared 5–7 days after nucleofection. Once iPS cell colonies appeared, they were allowed to grow in mTeSR1 media supplemented with a cocktail mix of 400 nM sodium butyrate and 100 μg ml−1 ascorbic acid. Once colonies were established, ReLeSR (5872, StemCell Tech) was used to select colonies. Standard timeline for reprogramming and expansion was 2–3 weeks for optimal conditions. A summary of iPS cell lines generated from patients with DMD is shown in Supplementary Table 2.
Cell culture
All iPS cell lines were cultured in mTeSR1 medium on Matrigel-coated flasks in an incubator at 37 °C at 5% CO2 until 60–80% confluency was reached, at which point cells were split using ReLeSR.
ASO design
All ASO molecules used were synthesized (100-nmol scale) at Integrated DNA Technologies with 2′-O-methyl modifications (Supplementary Table 3). Troponin-targeted ASOs were designed to overlap either the AUG translation start site, an early splice donor site or an early splice acceptor site based on the TNNT2 transcript structure annotated in GENCODE Release 43. For TNNT2-knockdown experiments, we synthesized a scrambled control based on the AUG-targeting ASO in which the sequence was randomly scrambled but nucleotide composition retained. We also synthesized a mismatched control based on the AUG-targeting ASO containing four nucleotide changes evenly spaced across the sequence (GC content retained). For DMD experiments with existing therapeutics, we synthesized fully modified 2′-O-methyl RNAs matching the sequences of the FDA approved and Phase II clinical trial compounds. As a negative control for these experiments, we synthesized an ASO with no complementarity to the human genome. For DMD experiments with personalized ASOs, we designed two distinct ASOs targeting sequences that overlap the intronic single-nucleotide variant detected in patients 2a and 2b. For each personalized ASO, we designed a distinct scrambled control in which the ASO sequence was randomly scrambled but nucleotide composition retained. In addition, we designed distinct mismatched controls containing four nucleotide changes evenly spaced across the ASO sequences (GC content retained). The sequences of all ASOs used in this study are shown in Supplementary Table 3.
ASO delivery
ASOs were added at a final concentration of 10 µM, which was introduced with the transfection reagent TransIT-TKO (MIR2250, Mirus). For example, TransIT-TKO was added to Opti-MEM (31985062, Thermo Fisher Scientific) and mixed gently by pipetting up and down. For every 100 µl of the Opti-MEM-ASO mix, 3 µl of TransIT-TKO was used. The ASOs were then added to the Opti-MEM-TransIT-TKO mix at the desired concentration and mixed gently by pipetting and incubated at room temperature for 45 min. After 45 min, the ASO transfection mix was added dropwise and gently shaken to evenly distribute the ASOs. ASOs were added to cardiac organoids 14–22 days after the start of differentiation, and media were changed 48 h later.
iPS cell–cardiac organoid differentiation
To differentiate iPS cells into cardiac organoids, cells were dissociated using Accutase (A1110501, Thermo Fisher Scientific) for embryoid body formation. After dissociation, cells were centrifuged at 300g for 3 min and resuspended in mTeSR1 medium (85850, StemCell Tech) containing 10 µM ROCK inhibitor (Y-27632 dihydrochloride; 1254, Tocris). iPS cells were counted and seeded at 5,000 cells per well in a 96-well ultra-low binding, U-shaped-bottom microplate (4515, Corning) or 5,000 cells per microwell when using the AggreWell 800 plates (34825, StemCell Tech). The plate was centrifuged at 100g for 3 min and placed in an incubator at 37 °C at 5% CO2. After 3–5 days, embryoid bodies were ready for cardiac organoid differentiation. Embryoid bodies have round and smooth edges when they are ready for differentiation. The cardiac differentiation protocol was based on a previously reported small-molecule differentiation method with modifications for use in cardiac organoid differentiation43,44,45. Cardiac differentiation was started by the addition of cardiac differentiation media (CDM; RPMI 1640/B-27; 11875119, A1895601, Fisher), minus insulin containing CHIR99021 (72054, StemCell Tech) at a final concentration of 6 µM. After 2 days, media were changed to CDM, minus insulin containing IWP2 (3533, Tocris) at a final concentration of 5 µM for 2 days. After 2 days, media were changed to CDM, minus insulin for an additional 2 days followed by the addition of maintenance media (RPMI 1640/B27 with insulin; 17504044, Fisher). Media were changed every 2–4 days with maintenance media for the duration of the experiment. The alternative cardiac organoid differentiation protocol was performed as previously described46.
Immunofluorescence
Cardiac organoids were fixed in 4% paraformaldehyde solution (252549, Sigma) overnight at 4 °C. Fixation was followed by washes in 1X PBS (D8537, Sigma) and permeabilization using 1X PBS with 0.2% Triton X-100 (X100, Sigma) for 30 min at room temperature. After permeabilization, blocking was performed overnight at 4 °C in blocking solution (1X PBS, 5% FBS (10082147, Thermo Fisher Scientific), 0.2% Triton X-100 and 2.5% BSA (A9418, Sigma)). The next day, cardiac organoids were washed 3 × 3 min in 1X PBS containing 0.1% BSA followed by the addition of primary antibody solution for 2–3 days at 4 °C. Primary antibody exposure was followed by 5 × 3 min washes and incubation with secondary antibodies for 24 h at 4 °C in the dark. After secondary antibody incubation, cardiac organoids were washed 5 × 3 min in 1X PBS. After secondary incubation, DAPI (AB228549, Abcam) was added at 1:1,000 for 15 min at room temperature, followed by three washes with 1X PBS. Antibody solution was 1:10 of blocking solution to 1X PBS. Primary antibodies were used at 1:100 dilutions (see Supplementary Table 4). Secondary antibodies (goat anti-rabbit Alexa Fluor 488 and goat anti-mouse Alexa Fluor 594; A11034 and A11032, Fisher) were used at 1:400. After immunostaining, organoids were incubated with fructose–glycerol clearing solution (60% glycerol and 2.5 M fructose; 2 h at room temperature followed by overnight at 4 °C) to optically clear organoids before imaging. Organoids were imaged using the Nikon W1 spinning disk confocal microscope.
Calcium imaging
Cardiac organoids were loaded with 10 µM Fluo-4 AM (F14217, Thermo Fisher Scientific) with 1.25 mM probenecid (P36400, Fisher) and 0.02% pluronic F-127 (P6867, Fisher) added directly to the maintenance media (RPMI 1640/B27 with insulin) for 45 min at 37 °C. The cardiac organoids were washed, and maintenance media were added back and incubated at 37 °C for an additional 30 min before responses were measured using a fluorescence microscope (Keyence BZ-X810) using the FITC filter set. Quantification of cardiac contraction was performed using Musclemotion, a free open-source software for ImageJ.
Trilineage differentiation
Directed differentiation into all three germ layers (endoderm, mesoderm and ectoderm) was achieved using the STEMdiff Trilineage Differentiation Kit (05230, StemCell Technologies) following the manufacturer’s instructions. In brief, iPS cells were harvested using ReLeSR and single cells were generated by gentle pipetting up and down. This was followed by counting and seeding with recommended cell densities onto Matrigel-coated 24-well plates. Cells were maintained in lineage-specific medium with daily medium changes until day 5 for mesoderm and endoderm differentiation, and until day 7 for ectoderm differentiation. Immunofluorescence staining was performed to assess differentiation to the ectoderm (nestin and PAX6), endoderm (FOXA2 and SOX17) and mesoderm (Brachyury and NCAM).
Karyotyping
Genomic integrity was assessed by karyotyping with a standard 20 iPS cell analysis. Karyotype services were provided by the Children’s Mercy Kansas City Hospital Genetics laboratory. In addition, the hPSC Genetic Analysis Kit (07550, Stem Cell Technologies) was used to check chromosome abnormalities following the manufacturer’s instructions.
Cell viability
Cardiac organoid viability was determined using the CellTiter-Glo 3D Cell Viability Assay kit (G9682, Promega) following the manufacturer’s instructions.
Fluorescently labelling ASOs
ASOs were labelled using the Label IT Nucleic Acid Labeling Kit MFP488 (MIR7125, Mirus) following the manufacturer’s instructions. In brief, the labelling reaction was assembled according to instructions, and was incubated at 37 °C for 3 h followed by purification using G50 microspin purification columns and transfected into cardiac and brain organoids as described above.
Flow cytometry
Transfection efficiency was assessed by flow cytometry using fluorescently labelled ASOs. Three days after organoid transfection, organoids were dissociated using TrypLE Express (126040132, Thermo Fisher). Organoids were washed once with 1X PBS and then incubated in TrypLE solution at 37 °C for 30 min. Subsequently, organoids were dissociated into single cells by gentle pipetting. Cells were then centrifuged down to remove TrypLE and washed once with 1X PBS (300g for 3 min). Pellets were resuspended in 1X PBS supplemented with 2% FBS and 4 mM EDTA in a final volume of 300 µl and stained with 7AAD for 15 min at 4 °C in the dark. Cells were washed twice with PBS–FBS–EDTA and then analysed on a BD Fortessa X-20 flow cytometer. The fraction of ASO-containing cells was measured by gating on total nucleated cells (forward scatter/side scatter), live cells (7AAD negative) and then the percentage of fluorescence-positive cells in the FITC channel. An untransfected control was used to subtract baseline fluorescence intensity. Individual flow cytometry analyses are shown in Supplementary Fig. 12.
RT–PCR
Total RNA was extracted using an RNeasy Mini Kit (74104, Qiagen) according to the manufacturer’s instructions. RNA was treated with DNAse I, eluted in 50 µl RNase-free water and quantified using a Qubit-4 fluorometer (Invitrogen) or NanoDrop One (Thermo Scientific). A total of 1 µg RNA was reverse transcribed using the SuperScript III First-Strand Synthesis System for RT–PCR (18080-051, Invitrogen) according to the manufacturer’s instructions, and 2 µl was used in PCRs. Amplification of cDNA was carried out with Ex Taq DNA polymerase (RR001A, Takara). The sequences of all primers used in this study are shown in Supplementary Table 5. Uncropped agarose gel scans are shown in Supplementary Fig. 13.
Western blot
Cardiac organoids were prepared in RIPA lysis buffer (89900, Thermo Fisher), and supernatant was transferred to a fresh 1.5-ml tube. Total protein was quantified using the Pierce BCA Protein Assay Kit (23227, Thermo Fisher) following the manufacturer’s instructions. Thirty micrograms of total protein were resolved on a 3–8% Tris Acetate Criterion XT Precast gel (3450129, Bio-Rad) for DMD or a 4–15% Criterion TGX Precast gel (5671083, Bio-Rad) for cardiac trophin T and vinculin. HiMark Pre-stained Protein ladder (Life Technologies) was used as a size standard for DMD or Precision Plus Protein Dual Color Standards (161-0374, Bio-Rad) for cardiac trophin T and vinculin. The gels were run at 50 V for 15 min followed by 150 V for 1.5 h and then transferred to a 0.4-μm polyvinylidene fluoride membrane (162-0262, Bio-Rad) at 30 V for 6 h at 4 °C. Membranes were blocked in blocking buffer (1X PBS with 0.05%Tween-20 and 5% skim milk) for 1 h at room temperature. An overnight incubation at 4 °C with primary antibody (DMD (1:100), cardiac trophin T (1:200) and viniculin (1:5,000)) was carried out followed by three washes in 1X PBS–0.05% Tween-20 (PBS-T). The membranes were then incubated with corresponding secondary antibody (1:5,000), either goat anti-mouse IRDye800CW (926-32210, LI-COR) or goat anti-rabbit IRDye800CW (926-32211, LI-COR). Western blot results were visualized on an iBright 1500. Uncropped western blot scans are shown in Supplementary Fig. 13.
Mutation site verification
The genomic DNA from iPS cell lines were extracted using the Nucleospin Blood Kit (740951.50, Macherey-Nagel) and amplified. PCR samples were purified using the QIAquick PCR Purification Kit (28106, Qiagen). All product sizes were confirmed on a 2% agarose gel before Sanger sequencing (Genewiz/Azenta Life Sciences) and Amplicon-Ez Next Generation sequencing (Genewiz/Azenta Life Sciences).
Amplicon sequencing
For visualization, raw sequencing data were aligned to the human genome using STAR with default settings and GENCODE Release 43 as the reference transcriptome47. To quantify ASO-mediated exon skipping, sequencing reads containing exon 44–45 junctions or exon 44–54 junctions were enumerated and normalized to the total number of sequencing reads for the sample.
Plasmids: skeletal muscle differentiation
LV-TRE-WT human MyoD-dsRedExpress2 was a gift from C. Gersbach (Addgene plasmid #60628).
Lentivirus production: skeletal muscle differentiation
HEK293T cells (American Type Culture Collection) were seeded (1 × 106 cells per well) in a six-well dish and allowed to attach for 24 h. A transfection mixture of 8.25 µl TransIT LT-1 reagent (Mirus Bio) with psPAX2 vector DNA (11260, Addgene; 1,250 ng), pMD2.G vector DNA (11259, Addgene; 125 ng) and LV-TRE-WT human MyoD-T2A-dsRedExpress2 DNA (96918, Addgene; 1,250 ng) was mixed and the volume brought up with OptiMEM. The transfection mixture was allowed to incubate for 30 min at room temperature and added to the cells. Lentiviral supernatant was collected 48 h post-transfection.
iPS cell–skeletal muscle differentiation
Skeletal muscle differentiation was based on previous published methods19. iPS cells used for skeletal muscle differentiation were maintained in growth medium containing Dulbecco’s modified eagle’s medium (DMEM)/F12 (21041025, Thermo Fisher), 20% knockout serum replacement (10828028, Thermo Fisher), 1% MEM non-essential amino acids (11140050, Thermo Fisher), 2 mM Glutamax (35050079, Thermo Fisher), 100 mM β-mercaptoethanol (21985023, Thermo Fisher) and 10 ng ml−1 bFGF (78003.1, StemCell Tech) supplemented with 0.4 µM PD0325901, 1 µM CHIR99021, 5 µM Y-27632 and 2 µM SB431542. To generate stable iPS cell lines with integrated Dox-inducible MYOD1 transgene, iPS cells were infected with the MyoD lentivirus supplemented with 4 µg ml−1 polybrene (TR-1003-G, Sigma). Uninfected cells were removed by 3-day incubation with 2 µg ml−1 puromycin (AT1113803, Gibco) to obtain a pure population of transduced cells. Following selection, iPS cells were pooled and expanded in growth media with puromycin. To prime cells for differentiation, iPS cells were seeded on Matrigel-coated plates in iPS cell growth media without bFGF supplemented with ROCK inhibitor (Y-27632). The next day, the medium was changed to induction medium (DMEM and 15% FBS) containing 3 µg ml−1 doxycycline (D9891, Sigma) to induce MyoD transgene expression. Medium was changed 4 days later to Dox-containing differentiation media (low-glucose DMEM (36253, StemCell Tech), 5% horse serum (26050088, Thermo Fisher) and 3 µg ml−1 doxycycline) and changed every 2 days.
Cerebral organoid differentiation
Cerebral organoid differentiation was based on previous established protocols with slight modifications48. In brief, iPS cells were dissociated and centrifuged at 300g for 3 min. Cells were resuspended in mTeSR1 media plus 10 μM ROCK inhibitor and single celled by gentle pipetting. Ten thousand cells were plated in a low-attachment 96-well plate, followed by centrifugation at 100g for 3 min and placed in an incubator at 37 °C at 5% CO2. Medium was changed every other day for 6–7 days. On days 6–7, when embryoid bodies have smooth edges and begin to brighten, the medium was changed to neural induction media (DMEM/F12, 1:100 N2 supplement (17502048, Invitrogen), Glutamax, MEM-NEAA and 1 µg ml−1 heparin (H3149, Sigma)) and refed every other day. After 4–5 days, neuroepithelia became visible and aggregates were coated in a 1% diluted Matrigel–DMEM solution and incubated at 37 °C for at least 30 min to allow Matrigel polymerization. Differentiation media without vitamin A (1:1 mixture of DMEM/F12 and neurobasal (21103049, Invitrogen) containing 1:200 N2 supplement, 1:100 B27 supplement without vitamin A (12587010, Invitrogen), 3.5 µl l−1 2-mercaptoethanol, 1:4,000 insulin (I9278, Sigma), 1:100 Glutamax and 1:200 MEM-NEAA) were added and refed every other day. After 3–4 days, differentiation media were changed to differentiation media with vitamin A (17504044, Invitrogen) and changed every 3–4 days. Cerebral organoids were ready for downstream analysis after 2–3 weeks.
Ethics statement
The Institutional Review Board of Children’s Mercy Research Institute gave ethical approval for this work (studies #00002465 and #11120514). All methods were carried out in accordance with relevant guidelines and regulations.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.