Patient biospecimen procurement and processing
Tissue collection
Patients undergoing synchronous colorectal resection and metastasectomy at MSKCC were identified by chart review, and those who had signed pre-procedure informed consent to MSK IRB protocols 06-107, 12-245, 14-244 and 22-404 for biospecimen collection were selected for this study. No statistical method was used to pre-determine sample size. Freshly resected surgical tissue in surplus of clinical diagnostic requirements was processed into single-cell suspensions for scRNA-seq analysis and, where sufficient tissue was available, processed to generate organoids. Portions were also fixed in formalin and embedded in paraffin. Tissue was generally processed within 1âh of surgical resection. Archival formalin-fixed, paraffin-embedded (FFPE) clinical tissue blocks for immunostaining were identified by database search and chart review. Tissue processing and histopathological data interpretation were overseen by an expert gastrointestinal pathologist (J.S.). Where trios of normal colon, primary CRC and metastatic CRC were successfully collected, the patient was longitudinally tracked through their clinical course at MSK using MSK Darwin52, and tumour tissue surplus to diagnostic requirements was collected from any subsequent procedures.
Patient metadata
Clinical data, including baseline demographic data and previous treatments (Supplementary Table 1 and Supplementary Fig. 1), were abstracted through manual review of patient electronic medical records by board certified medical oncologists (M.L. and K.G.), collected as part of institutional review board approved protocols (MSK IRB, 14-244 and 22-404). The time to each treatment event was calculated from the date of diagnosis to allow for comparison across patients. Study data were collected and managed using REDCap electronic data capture tools hosted at MSKCC on secure central servers. 17 out of 31 patients had multiple metastatic sites at the time of surgery, and had >50% of tumour sites remaining after surgery. 17 out of 31 patients had early-onset CRC (age of diagnosis, <50âyears). Clinical MSK-IMPACT targeted exon sequencing was performed on tumour/normal tissue from 27 out of 31 patients and revealed expected mutations53 (Extended Data Fig. 1b). Consistent with the low percentage (<5%) of metastatic CRC that is mismatch repair deficient/microsatellite instability high, only one patient in our cohort had an microsatellite instability indeterminate tumour. Clinical data collection was censored on 30 September 30 2022.
Tissue processing
We collected 50â300âmg of freshly resected surgical tissue in 5âml of IGFF organoid medium (Advanced DMEM/F12 (AdDF12; Thermo Fisher Scientific), GlutaMAX (2âmM, Thermo Fisher Scientific), HEPES (10âmM, Thermo Fisher Scientific), N-acetyl-l-cysteine (1âmM, Sigma-Aldrich), B27 supplement with vitamin A (Thermo Fisher Scientific)) supplemented with primocin (100âμgâmlâ1, InvivoGen), plasmocin (50âμgâmlâ1, InvivoGen), penicillinâstreptomycin (100âμgâmlâ1, Thermo Fisher Scientific), Amphotericin B (2.5âμgâmlâ1, Cytiva), nystatin (250âUâmlâ1, Millipore Sigma). For primary and metastatic tumours, specimens were placed into a 15âcm Petri dish using sterile forceps and washed three times with DPBS (Thermo Fisher Scientific) supplemented with the above-described antibiotic cocktail, and minimally chopped with sharp sterile blades to enable transfer of tumour fragments using a pre-wet 25âml serological pipette.
Tumour fragments were transferred into a gentleMACS type C tube (Miltenyi) pre-filled with 5âml of IGFF medium supplemented with antibiotics, DNase I (100âUâmlâ1, Millipore Sigma) and a commercial cocktail of tissue digestion enzymes (Tumour Dissociation Kit, Miltenyi). Tumours were digested using the gentleMACS Octo Dissociator according to the manufacturerâs 37C_h_TDK_1 protocol for a maximum of 30âmin. Considering the heterogeneity of tissue specimens with respect to cell viability, immune infiltration, blood content, necrosis and calcification, the digestion state of the tumour fragments was assessed every 10âmin under an inverted microscope. Digestion was interrupted before 30âmin if at least 50% of the tumour material appeared broken into clusters of 1 to 10 cells. Next, cell cluster solutions were filtered through a 100âμm cell strainer, and washed three times with DPBS supplemented with antibiotics, with each centrifugation step performed at 100g for 3âmin at room temperature. The final cell suspension was filtered through a 100âμm cell strainer, washed and centrifuged for 5âmin at 500g and 4â°C.
Non-tumour tissue was transferred into a 50âml tube pre-filled with 25âml dissociation/chelation buffer (8âmM EDTA, 0.5âmM DTT, DNase I (100âUâmlâ1, Millipore Sigma)). Mucosal fragments were incubated with gentle rotation at 4â°C for a maximum of 30âmin. The dissociation state of the tissue fragments was assessed every 10âmin under an inverted microscope. Dissociation was interrupted before 30âmin if at least 30% of the mucosal material appeared broken into clusters of 1 to 5 colonic crypts. Next, the crypt solution was filtered through a 1âmm cell strainer (PluriSelect) to separate individual crypts or small crypt clusters from large chunks of undissociated mucosa. Dissociation was quenched using an equal volume of DPBS supplemented with antibiotics. At this point, the 1âmm filter was flipped and inverted into a fresh 50âml tube. Up to 25âml of DPBS supplemented with antibiotics was flashed through the inverted filter to recover the undissociated mucosal tissue. After manually shaking the suspension of mucosal tissue fragments (approximately 5 times), the collection of clusters of colonic crypts was reattempted as described above. Based on the iteration of filtration and manual agitation steps, up to three additional fractions of crypt suspensions were collected. Crypt suspensions were washed three times with DPBS supplemented with antibiotics, with each centrifugation step carried out at 100g for 3âmin at room temperature. Based on visual inspection under an inverted microscope, one or more crypt suspensions were selected for subsequent processing according to the size and integrity of the crypts, and either processed separately or pooled together if individual suspensions were assessed to have low crypt content.
For both tumour and normal tissue, if blood traces were visible under an inverted microscope, the cell pellet was resuspended in 1â5âml ACK lysis buffer (Lonza), according to the pellet size and incubated for 5âmin at room temperature. Quenching was performed with three volumes of DPBS supplemented with antibiotics, followed by an additional wash to remove ACK traces. The resulting cell pellet was further processed for either scRNA-seq, organoid generation or both. Tissue processing protocols were extensively and iteratively optimized to maximize retrieval of high quality (low mitochondrial and ribosomal content) viable single-cell suspensions for downstream analyses.
scRNA-seq
Cell suspensions were filtered through a 40 μm cell strainer and incubated in FACS buffer (10âmM HEPES, 0.1âmM EDTA, 0.1% FBS) with DAPI (1âμgâmlâ1, Thermo Fisher Scientific) and calcein AM (Invitrogen) for 5âmin on ice. Viable (calcein positive) cells were sorted using a 130âμm nozzle (SH800S SONY sorter) and collected in DPBS with 0.04% bovine serum albumin (BSA). scRNA-seq was performed on the Chromium instrument (10x Genomics) according to the 3â² RNA v3.1 user manual. In brief, FACS-sorted cells were washed once with DPBS containing 0.04% BSA and resuspended to a final concentration of 700â1,300 cells per microlitre. Cell viability was above 80%, as confirmed with 0.2% (w/v) Trypan Blue staining (Countess II). Cells were captured in droplets and subjected to reverse transcription and cell barcoding; emulsions were then broken and cDNA was purified using Dynabeads MyOne SILANE followed by PCR amplification according to the manual instructions. Up to 10,000 cells were targeted for each sample. Final libraries were sequenced on the Illumina NovaSeq S4 platform (R1, 28 cycles; i7, 8 cycles; R2, 90 cycles).
Organoid generation and culture
Primary and metastatic CRC and normal colon organoid lines were established as previously described10,43,54. Cells processed as described above were centrifuged at 600g for 5âmin at 4â°C and resuspended at 2,000 cells per 40âμl of Matrigel. After Matrigel domes solidified at 37â°C, HISC medium supplemented with Y-27632 was added to the wells. Organoids were passaged every 7â10âdays, and were considered established after three passages. For non-tumour organoid culture, HISC medium was supplemented with human R-spondin 1 (1âμgâmlâ1; Peprotech) and NGS-WNT (0.5âM, ImmunePrecise N000). The medium was changed every 3â4âdays. Organoid lines were expanded and early-passage stock vials were cryopreserved in liquid nitrogen.
For validation, organoids underwent targeted exome sequencing by MSK-IMPACT53 and key oncogenic genomic alterations were identified by OncoKB55 (see below). Diagnostic tissue from originating tumours was sequenced to confirm that these alterations were conserved in each derived organoid line. Organoids were verified on the basis of short tandem repeats at the time of establishment and before every experiment, and were routinely tested for mycoplasma contamination (MycoALERT PLUS detection kit, Lonza).
MSK-IMPACT
Tumour and organoid targeted exon sequencing was performed using MSK-IMPACT53. The OncoKB precision oncology knowledgebase, an FDA-recognized human genetic variant database curated by experts at MSK55, was used to distinguish between oncogenic alterations (presumed drivers) and variants of unknown significance (presumed passengers). Only somatic alterations labelled as oncogenic, likely oncogenic or predicted oncogenic by OncoKB were included for analysis. The MSK-IMPACT data analysis pipeline is available at GitHub (https://github.com/rhshah/IMPACT-Pipeline). Genomic alterations were annotated with information from OncoKB using the OncoKB annotator tool (https://github.com/oncokb/oncokb-annotator).
FACETS
Copy-number alterations in solid tumours were computed from MSK-IMPACT using the FACETS (Fraction and Allele-Specific Copy Number Estimates from Tumour Sequencing) algorithm56, which provides allele-specific copy-number estimates at the level of both gene and chromosome arm. FACETS was also used to generate purity-corrected segmentation files, for detection of whole-genome duplication events, to infer the clonality of somatic mutations, to assess arm-level copy-number changes and to generate mutant allele copy-number estimates.
Computational data analysis
scRNA-seq data pre-processing
Alignment of sequencing reads
All scRNA-seq datasets were pre-processed as follows: FASTQ files from patient samples were processed with the SEQC (v.2.7.0) pipeline57 using the hg38 human genome reference, default parameters and platform set to 10x Genomics v3 3â² scRNA-seq kit. The SEQC (v.2.7.0) pipeline performs read demultiplexing, alignment and unique molecular identifier (UMI) and cell barcode correction, producing a preliminary count matrix of cells by unique transcripts. By default, the pipeline will remove putative empty droplets and poor-quality cells based on (1) the total number of transcripts per cell (cell library size); (2) the average number of reads per molecule (cell coverage); (3) mitochondrial RNA content; and (4) the ratio of the number of unique genes to library size (cell library complexity). However, due to the sensitivity of the colorectal epithelium to dissociation, we observed increased indicators of cell stress, apoptosis and droplet contamination in many samples, including high mitochondrial and ambient RNA expression, which can obscure statistical inference from meaningful biological gene expression. As such, typical ad hoc cell filtering based on identifying a steep dropoff in the number of transcripts per droplet (that is, a deviation leading to a âplateauâ in ambient RNA levels), could impair the extraction of meaningful biology. We therefore sought to systematically evaluate and correct for ambient RNA expression and filter for real single cells using CellBender (v.0.1.0)58 as described below.
CellBender to subtract ambient RNA
CellBender (v.0.1.0)58 is an unsupervised method for removing ambient RNA from scRNA-seq data. It first infers levels of ambient RNA and rates of barcode swapping per gene and droplet, respectively, from an unfiltered cell-by-gene count matrix. This probabilistic model is then used to generate a denoised (that is, ambient RNA-corrected) count matrix, as well as the probability that each droplet contains a cell, which can be used for calling real cells. We ran CellBender (v.0.1.0) on the unfiltered count matrix of each sample produced by SEQC (v.2.7.0) with the following parameters: (1) set the expected number of cells as the number of cells loaded into each 10x Chromium lane per sample (typically 5,000â10,000 cells); (2) set total droplets used to estimate ambient background RNA to 30,000; and (3) set training epochs to 100. We used the denoised count matrix produced by CellBender (v.0.1.0) for all subsequent analyses.
Removal of low-quality cells
On the basis of CellBender-corrected expression counts, we sought to identify and filter out low-quality cells from downstream analysis. As our study focuses on epithelial cells, which are known to be more sensitive to single-cell dissociation than other cell types, we paid special attention to droplet quality by performing three filtering steps:
Step 1: remove all droplets with posterior probability of containing cells â¦0.5 according to CellBender (v.0.1.0). This lenient filtering ensured that no biologically relevant cells were removed, at the cost of retaining some cells with worse technical characteristics. Step 2: remove droplets with <200 total counts, <200 total genes expressed or of which the libraries comprised >50% mitochondrial RNA. Step 3: Iterative rounds of clustering and filtering to remove low-quality or apoptotic cells that group together to create unstructured âjunkâ clusters. We carried out this filtration by combining count matrices from all patients for each sample type (non-tumour, primary tumour and metastasis), clustering cells using PhenoGraph11 (kâ=â20), and studying the covariance structure of highly expressed genes within each cluster. We reasoned that highly expressed genes are not co-regulated in cells undergoing apoptosis, nor in droplets containing ambient RNA, motivating the removal of droplets lacking meaningful covariance structure. We repeated the process of clustering and filtering until only cells residing in structured clusters remained, after which all datasets were combined.
Cells passing all of the above criteria were retained for downstream analysis.
scRNA-seq data analysis
Data normalization and dimensionality reduction
Raw count matrices were normalized to the median library size and log-transformed with a base of e and pseudocount of 0.1. We then selected highly variable genes (HVGs) using the highly_variable_genes function in Scanpy (v.1.9.1) and flavourâ=âseurat_v3 (we chose binsâ=â40). We kept the top 50 genes within each bin, for a total of 2,000 HVGs. Moreover, for all datasets (besides the fetal colon dataset), we included genes with known relevance to normal colonic cell types (41 genes) and cell states associated with inflammatory disease, injury of the colon, and regulation of REST and EMT (56 genes) (a complete list of genes is provided in Supplementary Table 7). Going forward, we will denote the 2,097 genes including both HVGs and manual additions as HVGs. Next, we performed PCA of log-normalized matrices using only HVGs and retained the number of principal components (PCs) that explain 75% of variance (112 PCs). For all datasets as well, we chose a number of PCs explaining 75% of variance.
Data visualization
For all two-dimensional embeddings, we used the Scanpy (v.1.9.1) neighbours function to compute a k-nearest-neighbour graph on the PCs based on Euclidean distance and kâ=â30. To visualize the global CRC cell atlas (Extended Data Fig. 2b,e,h,i), non-tumour epithelium (Extended Data Fig. 2f) and human fetal gut cell atlas (Fig. 2c), we generated projections using the UMAP implementation in Scanpy (v.1.9.1), with min_distâ=â0.3â0.5 and init_posâ=âpaga. To visualize epithelial cell subsets including all untreated epithelial cells (Extended Data Fig. 3a,c), all tumour cells (Extended Data Fig. 5eâj) and patient KG146 cells (Fig. 2a and Extended Data Fig. 9b,c), we used force-directed layouts, which provide a more intuitive representation of cell state transitions and the local relationships between subpopulations, using Scanpy (v.1.9.1) with the ForceAtlas2 layout and init_pos = paga. The Python package matplotlib (v.3.6.0) was used to produce all plots.
Gene expression denoising and imputation
We applied MAGIC (v.3.0.0) imputation59 to normalized, log-transformed count matrices to denoise and recover missing transcript counts due to dropout. Imputation was performed using conservative parameters (tâ=â3, kaâ=â5, kâ=â15). Imputed values are used for visualization of gene expression or gene signature expression (described in the main text and figure legends where used), as well as for analysing mixed-lineage gene correlations in untreated patient tumours (see the âGene correlations in normal intestine and untreated tumourâ section).
Gene signature scores
To generate all gene signature scores in our study, we used the Scanpy (v.1.9.1) score_genes function, which calculates the mean expression of genes of interest subtracted by the mean expression of a random, expression-matched set of reference genes. To account for expression-level differences across genes within signatures, we provided z-normalized expression data as the input for this function.
Cell annotation
Partitioning cells into epithelial, stromal and immune compartments
We clustered the dataset of all cells using PhenoGraph (v.1.5.7) with the Louvain algorithm (kâ=â45) on the PCs obtained above. To ensure robustness to the choice of k, we repeated PhenoGraph (v.1.5.7) clustering for all values of k between 20 and 100 in increments of 5 and calculated the adjusted Rand index between each pair of clusterings. We chose the value kâ=â45, which, within small variation, generated a Rand indexâ>â0.9, indicating that cell assignments to clusters remain mostly unchanged and are therefore robust to the choice of k.
We next partitioned clusters into epithelial, stromal and immune compartments based on marker gene expression (Extended Data Fig. 3a,b). Specifically, we used the score_genes function in Scanpy (v.1.9.1)60 to score expression of compartment-specific gene signatures from ref. 61, similar to the strategy used in that study (signatures for each compartment are shown in Supplementary Table 7). Each cluster was assigned to the compartment with the maximal score.
Analysis of the epithelial compartment
We filtered the epithelial compartment to discard any remaining low-quality cells, by removing the lowest modes in the distributions of log-transformed library-size, log-transformed number of genes expressed and the fraction of mitochondrial RNA, resulting in 67,534 epithelial cells. We chose to assign thresholds separately for each compartment due to their different sensitivities to dissociation and sample preparation, as well as inherent biological differences between compartments, for example, tumour cells often have very large library sizes compared to immune or stromal cells.
Within the epithelial compartment, we then recomputed HVGs (2,097 HVGs), re-performed PCA (210 PCs, 75% variance explained) and clustered cells with PhenoGraph (v.1.5.7) (kâ=â30) and removed the four remaining outlier clusters containing cells belonging to patients KG103, KG105 and KG66. These clusters were characterized by a very low library size; little block structure in their gene covariance matrices, primarily containing mitochondrial and ferroptosis-associated genes; and a strong overlap between cells originating from the patientsâ non-tumour, primary and metastatic samples. Consistent with our scRNA-seq data, we observed very aberrant mucosa in histological images of non-tumour samples for these patients, noting an association with previous disease conditions, which may explain the poor sample quality. Together, these observations suggested that the clusters probably represent highly stressed or dying, disease-associated cells that would not be informative to our study. After removing them, 47,437 cells remained; all downstream analysis of the epithelial compartment was performed on these cells.
Tumour cell identification using single-cell CNA calls
We identified cancer cells in the epithelial compartment (Extended Data Fig. 3câe) using the following criteria: (1) evidence of copy-number alterations (CNAs) compared with cells originating from non-tumour colon samples; and (2) clustering that is distinct from non-tumour epithelial cells.
We identified CNAs at the single-cell level using infercnvpy (v.0.4.0), a Python implementation of InferCNV12, using a sliding window of 200 genes and the default parameters. The mean expression of the reference diploid was determined using all available normal tumour-adjacent samples. We performed Leiden clustering on the inferred copy-number matrix and called cancer cell clusters if they had <25% normal tumour-adjacent cells and an average CNA score â§1âs.d. from the diploid mean. As a result, 3,102 cells derived from tumour samples with no CNA were reclassified as non-tumour epithelial cells (we performed cell type annotation on these cells in the section below), and 26,145 cells derived from tumour samples with CNA remained classified as tumour cells. We also produced an independent CNA estimate using the FACETS pipeline56 for samples from patients with targeted DNA panel sequencing data. In many cases, patient CNA estimates by FACETS were consistent with the most abundant single-cell CNA profiles computed with InferCNV for the same patients (an example is shown in Extended Data Fig. 3c,d).
Cell type annotation in the non-tumour epithelial compartment
To annotate epithelial cell types, we retained the subset of normal epithelial cells (21,297 cells) collected from the adjacent normal samples and those identified as normal from the tumour samples, computed a PC representation (249 PCs) of the resulting log-normalized count matrix, and clustered cells with PhenoGraph (v.1.5.7) using the Leiden option and kâ=â15 on the obtained PCs. We ensured robustness to the choice of k as described above. This process resulted in 48 clusters of cells, which were annotated into cell type based on two criteria: (1) similarity of mean z-normalized expression to that of canonical marker genes for major colon epithelial cell types (Extended Data Fig. 3g); and (2) GSEA of relevant cell type gene sets from the literature (Supplementary Table 2), based on differentially expressed genes (DEGs) in each cluster compared with the rest, computed using the R package MAST (v.1.16.0)62. GSEA was performed using the Python package gseapy (v.0.14.0)63 with 10,000 permutations and the default parameters. On the basis of both criteria, we manually annotated the clusters as ISC (2,580 cells), absorptive precursor (6,874 cells), enterocyte (2,115 cells), BEST4+ enterocyte (1,367 cells), secretory precursor (5,573 cells), goblet (1,751 cells), tuft (848 cells) and enteroendocrine (189 cells) (Extended Data Fig. 3fâj).
Comparison of normal ISCs and treatment-naive tumours
Creation of an ISC-specific gene signature
To determine ISC-specific marker genes, we performed differential expression analysis of ISC cells against all other non-tumour cells using MAST (v.1.16.0) on the normalized, log-transformed count matrix for non-tumour epithelial cells, and calculated gene rankings according to the âlog[P]âÃâlog[fold change] value for each gene (Supplementary Table 2). The final ISC gene signature consisted of the top 100 ranked DEGs (Pâ<â0.01 for all genes included; Supplementary Table 2). We calculated a gene signature score on the z-normalized expression of these genes using the score_genes function in Scanpy (v.1.9.1) (Fig. 3c).
PCA and annotation of PC1
We took a subset of the single-cell dataset containing all untreated normal and tumour epithelial cells (13,935 cells) and performed PCA of the log-normalized expression matrix. We focused on the first PC (PC1), which we note was responsible for 13.5% of the variance in the dataset (compared to 8.96% of variance described by PC2). To annotate PC1, we ordered all genes according to their feature loadings on PC1, excluding genes with zero-valued loadings which cannot be ordered. Using this ordering, we performed GSEA using the prerank function of the Python package gseapy (v.0.14.0) with relevant cell type gene sets from the literature13,21,61,64,65 (Supplementary Table 3) and the default parameters as inputs (Fig. 3d).
DEG and GSEA analysis between untreated tumour and ISC cells
We performed differential expression analysis of ISC cells against all untreated tumour cells using MAST (v.1.16.0) and GSEA using relevant cell type gene sets from the literature (Supplementary Table 3) as well as all Hallmark66 and KEGG67 gene sets (Extended Data Fig. 3f). GSEA was performed using the prerank function of the Python package gseapy (v.0.14.0) with 10,000 permutations and the default parameters (Supplementary Table 3).
Identification of ISC phenotypic admixture in treatment-naive tumours
To assess whether the cell type promiscuity observed at the cluster level (Extended Data Fig. 3e) is also present in individual cells, we used MAST (v.1.16.0)62 to compute DEGs enriched in ISCs, enterocytes and goblet cells in the non-tumour epithelial dataset; ranked each DEG by âlog[P]âÃâlog[fold change] value; and used the top 300 genes as lineage markers (Supplementary Table 3). For enterocytes and goblet cells, DEG analysis was restricted to differentiated cell types (that is, excluding precursor cell types). This allowed genes shared between precursor and differentiated cell types of the same lineage to be recovered by DEG analysis. For example, the expression of enterocyte marker SLC26A3 in late-absorptive precursor cells lowered its observed differential expression in enterocytes compared with all other cells when absorptive precursors were included in DEG analysis, despite abundant SLC26A3 expression among enterocytes.
As tumour cells do not emulate the complete phenotype of normal differentiated cell types68, we restricted the 300 cell lineage markers from above to genes that are also abundantly expressed in more than 20% of primary tumour or metastasis cells. We consider a lineage marker to be abundantly expressed in a cell if its normalized expression in that cell was greater than or equal to the bottom quartile of expression from the lineage for which it is a marker. To ensure the specificity of markers, we also removed any gene that is abundantly expressed in other lineages. Finally, we determined the fraction of the remaining lineage markers that are abundantly expressed in each normal cell and treatment-naive tumour cell. The distributions for each cell type and tumour type are visualized using the sns.kdeplot function in Python Seaborn (v.0.11.2) (Extended Data Fig. 3hâj).
Gene correlations in normal intestine and untreated tumour
To understand whether the co-expression of conflicting cell type markers (Extended Data Fig. 3hâj) is associated with the loss of cell-type-specific gene regulation in tumours, we first computed Pearson correlations between all pairs of top ranking DEGs for ISCs, enterocytes and goblet cells as described above (300 genes total) using the imputed expression matrix for (1) all non-tumour epithelial cells and (2) all treatment-naive tumour cells (Extended Data Fig. 3g). To account for heterogeneity in gene regulation across tumours, correlations were first computed for each patient tumour separately, resulting in 12 correlation matrices, and averaged. In this way, the observed strong correlations correspond to those gene pairs with strong positive or negative correlations across multiple patient tumours, suggesting consistent gene dysregulation compared to the non-tumour setting.
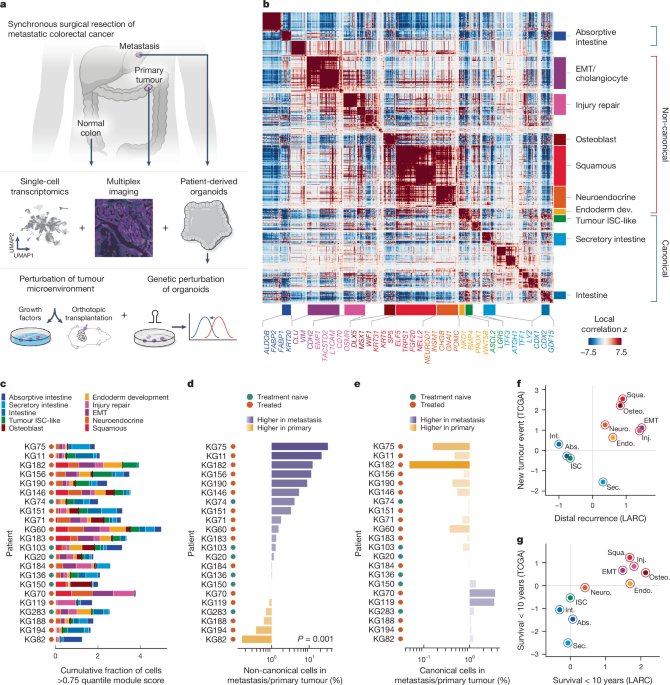
Identification of Hotspot gene modules in CRC tumour data
We used Hotspot (v.0.9.1)20, an algorithm for identifying context-specific gene modules within single-cell datasets given a user-provided metric for local cellâcell similarity, to identify shared context-specific gene modules in our patient tumour dataset. Hotspot evaluates the pairwise local correlation between genes in local cell neighbourhoods within the cellâcell similarity (k-nearest neighbours (k-NN)) graph, identifying genes with high local autocorrelation. Importantly, the way significant local autocorrelation is detected is suited to scRNA-seq, and is resilient to issues such as gene drop-out in individual cells. The computed geneâgene affinity matrix is clustered to output a set of gene modules. In contrast to global measures of correlation (such as Pearsonâs correlation), which presume relationships between features are consistent across a single-cell dataset, Hotspotâs local correlation measures are computed on the local cell neighbourhoods in the k-NN graph.
While non-negative matrix factorization has been used to identify cancer gene programs21,22, it is a linear method that requires a consistent and complete decomposition of the entire dataset, and it can be sensitive to batch and other variation. By contrast, Hotspot is based on geneâgene covariance, which better represents sets of genes that work together towards common functions, and is robust to batch effects57 (and is therefore likely to be more robust to variation between tumours). Importantly, Hotspot gene modules are based on covariance that can be localized to a cell subpopulation or exhibit potentially nonlinear graded expression over regions of the manifold. In cancer in particular, heterogeneous cell states adapted to different patient tumour contexts, such as metastatic sites, are likely to correspond to differences in gene covariance. Hotspot is also well suited to handle both gene pleiotropy and rare populations, which have important roles in tumour contexts. We therefore chose Hotspot to characterize sources of inter- and intra-tumour phenotypic heterogeneity in our dataset alongside global, manifold-defining modules of genes.
To apply Hotspot, we first partitioned the data to only include tumour cells (26,145 cells), and used their top 2,097 HVGs (see the âData normalization and dimensionality reductionâ section above). After renormalization, we performed PCA and retained enough PCs to explain 75% of the variance (233 PCs). We then identified a subset of significantly autocorrelated features with respect to the PC latent space by running Hotspot using the depth-adjusted negative binomial (danb) observation model and 30 neighbours. The danb model69 comprises the background null distribution against which expression counts are normalized, and is used to avoid flagging genes as significant due to local autocorrelation in the library size of cells.
We retained 2,003 genes with a FDRâ<â0.01 for calculating local correlations and downstream clustering (see below). We used the create_modules function in Hotspot (default parameters except for minimum_gene_thresholdâ=â20 and core only = False) to obtain a preliminary set of 37 co-varying gene modules (gene assignments to modules are shown in Supplementary Table 4).
Hotspot module clustering
Hotspot clusters the geneâgene local correlation matrix into modules using an agglomerative hierarchical clustering procedure that, at each step, merges two genes/modules with the highest pairwise z-scored correlation. Once a merged module contains more genes than a minimum threshold, it is labelled and cannot be combined with other labelled modules. The process ends when the highest pairwise z-score between unmerged modules falls below a minimum value; at that point, all genes that are unlabelled are not assigned to any module.
In practice, we found that choosing a large value for the minimum gene threshold left too many genes unassigned and created modules that, due to their high numbers of genes, were difficult to interpret. Choosing too small of a threshold failed to merge some intercorrelated, biologically similar modules. We opted to use a lower threshold of 20 genes minimumâto err on the side of more manageable, smaller modulesâand then manually group modules with similar biological interpretations, after ensuring their genes were also correlated. This process is described in detail below.
Hotspot module grouping and annotation
All original 37 Hotspot modules were annotated manually based on known canonical markers for intestinal and non-intestinal cell types and from the literature (a subset of annotation genes is shown in Fig. 1b and Extended Data Fig. 5b,c, and all annotation genes are shown in Supplementary Table 4). Gene set over-representation analyses using gseapy (v.0.14.0) and the Gene Ontology Biological Process gene sets provided supporting evidence for our initial annotations, or suggested possible directions to investigate for modules that were difficult to annotate (module annotations and over-representation analysis for final module annotations are shown in Supplementary Table 4).
We focused on 23 out of the 37 modules (1,201 genes) representing meaningful biological gene programs and did not further explore 14 modules (722 genes) annotated as cell cycle/proliferation (2 modules), cell stress (4 modules), leukocyte (3 modules) or cilia (1 module), or else could not be interpreted (4 modules). We then manually grouped 19 modules into 6 groups after (1) arriving at the same biological interpretation for all modules within a group, and (2) ensuring the local correlations between genes of grouped modules were high on average (Extended Data Fig. 5a,b). These six grouped modules and four single modules resulted in ten final gene modules (Extended Data Fig. 5a,b and Supplementary Table 4).
Once the modules were annotated and grouped using the above-described strategies, we categorized them into two distinct categories:
-
(1)
Canonical: modules describing canonical intestinal cell types and processes such as epithelial differentiation, mucus production and small-molecule transport, which are critical for maintaining normal intestinal function.
-
(2)
Non-canonical: modules describing processes not typically seen in healthy intestine, such as keratinization, inflammatory response and wound healing.
Hotspot gene module scores
Hotspot modules scores were calculated for our tumour dataset using Hotspotâs calculate_module_scores function. In brief, Hotspot evaluates per-cell scores for each module using the following procedure: for all genes in a module, expression counts are first mean-centred using the danb null model and smoothed using the weighted average of their nearest neighbours. The background null distribution factors cell library size differences into account; thus, centring based on the null ensures that correlations are not impacted by library size differences. PCA is then performed on the resulting centred counts and the first PC values are used as the module scores for each cell. We used Hotspot module scores to visualize and summarize the patterns of module expression within cell groupings (for example, clusters), and to relate gene modules to gene signature scores for pre-existing gene sets as well as to sample metadata.
To plot groups of cells with top-scoring expression for a given module, we used the scanpy dotplot function (Extended Data Fig. 5b,c). To assign cells as top-scoring for a Hotspot module, we z-normalized all Hotspot module scores across cells, and then required a top-scoring cell to very specifically express a given Hotspot module and not express any others. To ensure this, we required top-scoring cells to have (1) a score of >1âs.d. above its mean module score, and (2) a score of <1âs.d. above the mean for all other modules (cells are assigned to 1 out of the 23 ungrouped modules on which we focus in Extended Data Fig. 5b and to 1 out of the 10 grouped modules in Extended Data Fig. 5c). We found these criteria to be more selective than, for example, the percentile scores used in other sections when identifying cells that are high for a given module.
Robustness of Hotspot modules
We evaluated the robustness of the Hotspot analysis to the number of HVGs used as input features, based on the consistency of gene autocorrelation, and the consistency of obtained modules. We also evaluated the robustness of the Hotspot modules to bootstrapping of cells.
Consistency of gene autocorrelation to number of HVGs: we evaluated whether the input cellâcell similarity matrix faithfully captures the structure of the data across input features (highly variable genes). Given an input gene set, Hotspot removes genes with low autocorrelation along the k-NN graph, ensuring that only genes that vary along the manifold in an informative manner are selected for module detection. To determine whether genes passing this criterion are robust to the number of selected HVGs, we recomputed modules with 1,000â5,000 HVGs in increments of 500 genes, keeping all other parameters constant. For each combination, we calculated the difference in Hotspot local autocorrelations for each gene against its autocorrelation score when we input 2,000 HVGs (the value used in this study) and visualized the average as a box plot (Supplementary Fig. 2). These differences are minimal across Hotspot runs for all HVG pairs (maximum difference of 0.07), suggesting that the cell similarity graph retains its structure regardless of how many HVGs are chosen.
Consistency of modules to number of HVGs: to verify that the set of genes comprising each module is robust to the number of features, we generated Hotspot modules for 1,500â2,500 HVGs in increments of 50 genes, with minimum_gene_threshold set to 20 and core_only set to false. We then calculated Pearson correlations between the module scores of each set of Hotspot modules and the set obtained with 2,000 HVGs, as used in this study. For each original Hotspot module obtained with 2,000 HVGs, we report the correlation of the module used in this study and the best-matching moduleâthe module that is most highly correlated with the original module in this study (Supplementary Fig. 2). In general, every set of gene features identified a subset of modules showing close correspondence to our final set of modules based on max correlation.
Robustness of Hotspot gene modules to cell downsampling: to determine whether Hotspot modules depend on the exact cells used as input, we used a bootstrapping-like approach. We randomly resampled our tumour dataset ten times, with 1â10% of cells removed (in increments of 1%) and with 10%, 15% or 20% of cells removed. For each resampling, we calculated new PCs and a new k-NN graph, and reran Hotspot with the same genes and identical parameters as in our main analysis. We then evaluated the similarity of each Hotspot analysis with our original analysis as the maximal difference in local correlations (that is, the difference between the highest and lowest local correlation values across the matrices being compared) (Supplementary Fig. 2). We used this approach because cells cannot be resampled with replacement from a k-NN graph.
Relationship of Hotspot results to global correlation: while Hotspot geneâgene correlation is only evaluated on cell subsets, the subsets are highly constrained to the neighbourhood structure of the k-NN graph, which is a strong structural feature of the data, and makes it unlikely that Hotspot will find spurious modules. Nevertheless, we expect that these modules will also result in detectable levels of global Pearson correlation. For visual comparison, we plotted pairwise Pearson gene correlation against z-scored pairwise Hotspot local correlation on the log-normalized expression matrix (Supplementary Fig. 2), revealing that the ranking of weak and noisy global signals detected by global correlation largely align with the robust signals sensitively detected by Hotspot. Averaging all pairwise correlations within grouped modules likewise reveals that correlation and global correlation are qualitatively similar (Supplementary Fig. 2).
Distribution of module expression among samples
To determine the distribution of cells with high expression of the ten Hotspot modules on a per-patient basis, we first labelled a given cell with a gene module if it had >0.75 quantile expression score for that module, then plotted the cumulative fraction of labelled cells for all modules in each tumour sample (Fig. 1c), or in pooled primary and metastatic samples per patient (Extended Data Fig. 6g). Cumulative fractions can exceed 1 as a cell can exhibit high expression of more than a single module.
We also visualized non-canonical or canonical module prevalence (Fig. 1d,e) as the log-ratio of labelled cell fractions in metastatic to patient-matched primary tumours. Canonical and non-canonical module classifications are described in the âHotspot module grouping and annotationâ section. Although cells can exhibit high expression of multiple canonical or multiple non-canonical modules, we found that they do not often express both canonical and non-canonical modules highly (Supplementary Fig. 2); thus, we labelled cells as non-canonical or canonical according to which classification they score highest (for example, a cell of which the maximum module score is squamous is labelled non-canonical).
We calculated significance values for these plots using the following strategy: For each binary labelling of cells (for example, non-canonical or not non-canonical), we randomly permuted the labels 1,000 times within each patient separately. For each random permutation, we calculated the log-ratios of positive cells (for example, non-canonical) in metastases to positive cells in primary tumours, as above. We then performed a rank-sum test comparing our original log-ratios to the combined log-ratios from the random permutations. The alternative hypothesis is that our sample distribution is greater than the null (for example, metastasis is higher).
Interpatient entropy of gene modules
We used entropy to evaluate the patient-specificity of each Hotspot module using the following procedure: (1) sample 357 cells per patient with replacement in our tumour dataset to ensure even cell distribution across patients. (2) For each module, determine the subset of high-scoring cells with module scores greater than 1âs.d. above the mean. (3) Compute the Shannon entropy of patient labels in the high-scoring subset of cells for each module using the SciPy (v.1.9.1) function scipy.stats.entropy. To calculate the Shannon entropy, we first built a k-NN graph with kâ=â60 on the multiscale space embedding of all epithelial cells; multiscale space was computed on the top 19 DCs (chosen by the knee-point of DC eigenvalues) using Palantir (v.1.2). Steps 1â3 were repeated 100 times before visualizing the entropy distribution with kernel density plots using the kdeplot function in Python Seaborn (v.0.11.2) (Extended Data Fig. 5d).
Association of gene modules with clinical covariates in bulk RNA-seq data
To test the association of each Hotspot modules with clinical covariates in bulk cohorts, we (1) ran single-sample GSEA on two bulk RNA-seq datasets, LARC and TCGA-COAD, collected from CRC patient primary tumours using the genes in our Hotspot modules as the input gene sets; and (2) for each dataset, tested the association of patientsâ clinical features with the enrichment scores of their tumour samples.
ssGSEA analysis
For LARC, we analysed 108 LARC tumour samples with available RNA-seq data32. Genes were retained if they had >1 count per million across more than 50% of the samples. The edgeR v.3.40.2 package70 was used for trimmed mean of M-values normalization and FPKM transformation and the org.Hs.eg.db v.3.16.0 package was used for gene annotation. Genes that mapped to multiple Ensembl IDs were removed. We then performed ssGSEA analysis using the hacksig v.0.1.2 R package30 and all Hotspot modules. In this cohort, 0.4% of patients have a survival status of alive and an overall survival (OS) follow-up time of <12 months (0% with OS follow-up of <6 months); 1.7% of patients have no distal recurrence and a disease-free survival (DFS) follow-up time <12 months (0% with DFS follow-up of <6 months).
For TCGA, we downloaded and analysed RNA-seq data for 445 tumour samples from the TCGA-COAD study31. RNA raw counts were retrieved using TCGAbiolinks (v.2.26.0)71 and genes with a count of 0 across all the samples were removed, as well as genes that had multiple associated gene symbols or no gene symbol. The VST transformation was performed using the DESeq2 (v.1.38.3) package72. Subsequently, ssGSEA analysis was conducted utilizing the R package GSVA (v.1.46.0)73. In this cohort, 13% of patients have a survival status of alive and an OS follow-up time of <12 months (7% with OS follow-up of <6 months); 0.9% of patients have no new tumour event and a DFS follow-up time of <12 months (0.7% with a DFS follow-up of <6 months).
Associations with clinical covariates
For binary clinical features, we separated the enrichment scores into two groups based on the status of the patient from which the bulk RNA-seq data were collected and compared the enrichment scores for each Hotspot module between the two groups using the MannâWhitney U-test (Fig. 1f,g and Extended Data Fig. 6hâj).
Survival analyses in TCGA-COAD cohort
For each Hotspot module, we collected two groups of samples from the TCGA-COAD cohort: (1) highly enriched samples with ssGSEA enrichment scores greater than 1âs.d. above the mean enrichment score among all samples, and (2) low-enriched samples with enrichment scores less than 1âs.d. below the mean. We then performed a log-rank test on DFS between these groups using the lifelines (v.0.27.4) package in Python74. We generated survival curves for all modules with significant results (Pâ<â0.05) (Extended Data Fig. 6k). Multivariate logistic regression models were used to evaluate associations between DFS, OS and module expression. Each sample was annotated as high or low for each signature based on an ssGSEA score >0.75âs.d. above or below the mean, respectively. Samples without the signature annotation were excluded from the analysis. Cox proportional hazards tests were used for multivariate analysis of DFS. We generated forest plots for all modules with significant results (Pâ<â0.05) (Supplementary Fig. 3). R packages survival (v.3.6-4) and survminer (v.0.4.9) were used for the survival analysis.
Delineation of canonical to non-canonical tumour axes across patients
To characterize trends in cancer progression, we analysed the four patients with a sufficient number of cells in non-canonical states for robust characterization, namely KG146 (3,351 cells), KG182 (935 cells), KG183 (1,203 cells) and KG150 (2,574 cells). We reprocessed each patient individually to most faithfully capture trends within each individual patient; data from primary tumour, synchronous metastatic tumour and metachronous metastatic tumour samples were pooled for each patient and processed as described in the âData normalization and dimensionality reductionâ section. We used DC analysis, which identifies the largest axes of nonlinear variation in the data and has been shown to effectively capture cell-state transitions in scRNA-seq data75. DCs were independently computed for each patient to separately compute potential per-patient routes of tumour progression and to avoid artificially imposing the trends from patients with large samples on patients with smaller samples.
We computed diffusion maps (kâ=â30 nearest neighbours) for each patient and retained a subset of DCs based on the eigengap of the ranked componentsâ eigenvalues (KG146, 4; KG182, 6; KG183, 8; KG150, 4 DCs). The strongest DCs appear to define a continuum from canonical to non-canonical fates. Thus, we ranked the DCs of each patient by the difference between the average Spearman correlation of a given DC with (1) all non-canonical modules and (2) all canonical modules; the greatest difference between the two averages defines an axis for canonical to non-canonical transformation (Fig. 2b and Extended Data Fig. 7a). For KG146, KG182 and KG150, the first DC was selected, while the fourth DC was selected for patient KG183. The consistent and independent selection of the first DC as the canonical to non-canonical transition in 3 of 4 patients supports the importance of this axis as one of the strongest signals in the data. We note that KG183 had fewer non-canonical cells than the other patients, probably explaining why this transition was not the top DC for this patient. We used the progressive nature of our samples (normal to primary to metastasis) to reason thatâas normal included only canonical, and metastasis contained the largest fraction of non-canonical cellsâthis axis indeed represents a cell-state progression.
Visualization of module trends
We used generalized additive models (GAMs) with cubic splines as smoothing functions as in Palantir (v.1.2)42 to analyse module score trends along DC axes (Fig. 2b and Extended Data Fig. 7a). GAMs increase robustness and reduce sensitivity to density differences, and cubic splines are effective in capturing non-linear relationships. We fitted trends for a module score using a regression model on the DC values (x axis) and module score values (y axis or colour intensity). The resulting smoothed trend was derived by dividing the data into 500 equally sized bins along the DCs and predicting the module score at each bin using the regression fit. We visualized module score trends from the 20th percentile value (white) to the maximum value (highest saturation) (Fig. 2b and Extended Data Fig. 7a).
Derivation of a human fetal colon progenitor gene signature
We used a fetal gut cell atlas containing scRNA-seq data from dissected human embryos aged 6.1 to 17 weeks, which captures the development of human intestinal cells from a fetal progenitor state to differentiated crypts40. We downloaded a raw H5AD file from the authors containing all epithelial cells from fetal donors and limited our analysis to 8,408 cells originating from the large intestine of first and second trimester samples.
Data reprocessing and DEG analysis
As large intestine cells represent a minority of the fetal gut cell atlas (8,408 of 52,184 total cells), we partitioned and reprocessed the dataset to focus our analysis on these cells. We ran HVG selection (2,000 HVGs), PCA (167 PCs explaining 75% variance) and UMAP projection (min_distâ=â0.5) using scanpy as described in the âData normalization and dimensionality reductionâ and âData visualizationâ sections. UMAP projection revealed that the week 11.1âsample separated from all others. Cells from this sample were characterized by heat-shock genes DNAJB1, HSP90AA1, HSPE1, HSPA8 and HSPA1A among the top 10 DEGs (compared to all other samples, by MAST analysis) indicating cell stress, so it was removed, resulting in 7,984 cells. In the remaining dataset, we retained the authorsâ original cell type annotations, but merged the enteroendocrine subtypes (M/X, D, β, L, N, K, I and enterochromaffin cells) into one group.
We found that the first trimester samples consist predominantly of progenitor cells; proximal progenitor, distal progenitor and stem cells comprise 88% of all cells. By contrast, second-trimester samples consist of mature colon mucosal cell types exclusively, and exhibit strong expression of LGR5, TFF3, SLC26A3, NEUROD1 and POU2F3 (corresponding to ISCs, goblet cells, mature enterocytes, enteroendocrine and tuft cells, respectively). We therefore concluded that the separation between the first- and second-trimester samples captures the distinction between progenitor-like cell types and colonic crypts.
To determine marker genes specific to the first trimester cell population, we performed a differential expression analysis of first versus second trimester cells using MAST (v.1.16.0) on the normalized, log-transformed count matrix and identified 173 DEGs with log[FC]â>â2 and adjusted Pâ<â1âÃâ10â5. Earlier cells are more proliferative, so we removed genes related to cell cycle or proliferation from our first-trimester gene list. Specifically, we calculated the Pearson correlation of all first trimester DEGs with 445 genes belonging to the Reactome âCell Cycle, Mitoticâ and âCell Cycle, G1-G1/S Phaseâ gene sets and the Hallmark âCell Cycle, G2M Checkpointâ gene set76. We removed 60 genes with a correlation greater than 0.25 for at least one gene belonging to the gene sets. Our fetal gene signature comprises the remaining 113 genes (Supplementary Table 5).
Comparison with existing dedifferentiation signatures
We compared our 113-gene fetal signature to previously published dedifferentiation signatures23,34,38,39. For each pair of signatures, we calculated the Jaccard index (number of genes shared between signatures divided by total number of genes in both signatures), demonstrating that existing signatures are clearly distinct from our fetal signature and lack consensus (Extended Data Fig. 7b). We also determined how many of the 14 core fetal signature genes (see the next section) are present in each dedifferentiation signature, normalized to the total number of genes in that signature.
For each of the three major progenitor cell populations in first trimester dataset, and the five major populations in the second trimester dataset described above, we calculated the average score of the various signatures (see the âGene signature scoresâ section), finding that our fetal signature is clearly enriched in first trimester and depleted in second trimester populations, whereas other signatures lack coherent enrichment trends (Extended Data Fig. 7c).
Mapping fetal signature along tumour progression axis
For each patient, KG146, KG182, KG183 and KG150, we calculated gene set scores using the scanpy function score_genes and the list of 113 genes in our fetal signature. We determined which genes in our fetal signature are correlated (Pearson râ>â0.5) with the fetal signature score trend along the major diffusion component, as in the âIdentification of fetal-state-associated transcription factorsâ section below (Extended Data Fig. 7d). Among the 113 genes of the fetal signature, 88 genes are strongly correlated in at least one patient; 59 in at least two patients; and 37 in at least three patients, with a large number of genes correlated for each patient (56 in KG146, 51 in KG182, 29 in KG150 and 62 in KG183). Moreover,14 are strongly correlated with the signature score in every one of the four patients, forming a âcore signatureâ of candidates for driving fetal state reversion in patient tumours. We calculated a gene set score using these 14 core genes, as well and a significance value was calculated for its distribution among samples as in âDistribution of module expression among samplesâ above (Extended Data Fig. 7e).
KaplanâMeier analyses of fetal signature in bulk data
For all of the samples in the LARC and TCGA-COAD cohorts, we calculated ssGSEA enrichment scores as described in the âssGSEA analysisâ section using our fetal gene signature as input. For each cohort, we then collected (1) highly enriched samples with ssGSEA enrichment scores >1âs.d. above the mean enrichment score among all samples; and (2) lowly enriched samples with enrichment scores <1 s.d. below the mean. We performed a log-rank test on DFS between these groups using the Python lifelines (v.0.27.4) package (Extended Data Fig. 7g,h).
Pseudo-ordering of cell states by module overlap
Although diffusion component analysis orders patient cells along transitions from canonical to non-canonical fates in a reproducible and unbiased way, it is less effective in patients whose tumours have few cells in non-canonical states. As an alternative, we consider the observation that in most cell-state trajectories, gene modules tend to be co-expressed in the same cells if they define pairs of sequential cell states. Using this logic, the existence of a substantial fraction of cells co-expressing two distinct gene modules can be used to suggest a pseudo-ordering of these states, and a transition between them. A key feature of Hotspot is that a cell can co-express multiple modules (Fig. 1b and Supplementary Fig. 2), making it possible to examine cells occupying mixed states. To ensure that we only consider robust module expression, we assigned a cell to a module if it expresses module genes above the 75th percentile. For all pairs of Hotspot modules, we computed the fraction of modules that are co-expressed in the same cell, and we aggregated across patients due to the sparsity of some cell states in any given patient. This analysis reveals a progression (block diagonal) that agrees with DC analysis of the four patients with non-canonical states (KG146, KG182, KG183 and KG150) and is consistent across these four patients (Fig. 2d), the full cohort with these four patients removed (Fig. 2e), and five patients from an independent dataset (Fig. 2f).
Replication in an independent CRC scRNA-seq dataset
The scRNA-seq dataset from ref. 28 consists of matched primary tumour and liver metastasis samples for each of five patients who received multiple cycles of chemotherapy. We downloaded GEO accession GSM7058755 (colorectal cancer, nonimmune cells) comprising all tumour and stromal cells from these patients. We first filtered all cells with >40% mitochondrial UMIs or <1,000 UMIs, leaving 23,341 cells. We normalized data and classified epithelial cells using the same methods described for our data in the âscRNA-seq data analysisâ and âCell annotationâ sections; in brief, each PhenoGraph (v.1.5.7) (kâ=â30) cell cluster was classified according to the highest average score across all cells for a broad panel of stromal and epithelial genes. Only these filtered epithelial cells were used in downstream analyses.
Hotspot module analyses differ between our dataset and the ref. 28 dataset in two ways. First, gene module scores in our dataset are calculated using the k-NN graph that was generated to run Hotspot (see the âHotspot gene module scoresâ section), whereas the ref. 28 dataset requires its own k-NN graph, rendering the results impossible to compare. Instead, we calculated gene set scores using the scanpy function score_genes and the list of genes for each Hotspot module. Second, in our dataset, we used 0.75 quantile score thresholds to quantify per-patient cell abundances of Hotspot modules (Fig. 1b). As these are relative thresholds, repeating the analysis in the ref. 28 dataset would not account for the possibility that module genes are expressed at lower levels in ref. 28 compared with our tumour dataset. For this reason, we chose to construct thresholds which specifically reflect the level of expression in our dataset where we know these modules are expressed. To do so, we combined the two datasets and renormalized (median library size normalization and log-scale expression) to the same level of expression. Gene set scores were calculated on the joined, renormalized datasets using score_genes and the 0.75 quantile cut-offs were based only on the cells from our dataset, so that thresholds would reflect the level of expression in our dataset and be applicable to the data from ref. 28. These thresholds were used in Extended Data Fig. 6g.
Palantir pseudotime and branch calculations
The four patients whose tumours include the most cells with non-canonical fates (KG146, KG182, KG150 and KG183) span the spectrum of progression, and all contain both squamous and neuroendocrine states. We used Palantir (v.1.2)42 to investigate these two fates and the genes underlying their respective fate transitions. As an input, Palantir requires an initial state, and as the output, it computes terminal fates and provides a cell-fate map that assigns a probability for each cell to differentiate into each terminal fate. Palantir also outputs a pseudotime alignment of cells from the initial to each of the terminal states and, therefore, by combining pseudotime and fate probability for each cell, it can provide branching gene trends leading to each terminal fate (by weighing the contribution of each cell to the gene trend based on the fate probability). Palantir was run separately on the tumour datasets of patients KG146, KG182 and KG150, excluding KG183 because of an insufficient number of non-canonical cells in this patient. We selected cells with the highest imputed expression of LGR5 as the initial state, motivated by their identification as cells of origin in CRC studies. Notably, Palantir has been shown to be robust to the exact choice of starting cell. Running Palantir with 500 waypoints and an eigengap-based number of DCs (6 for KG146, 4 for KG182 and 8 for KG150) yielded distinct branching trajectories from the LGR5+ state to two CDX2â (non-intestinal) terminal cells in all three patients. We disregarded three additional branches in KG146 and KG150 that probably represent canonical-state trajectories, as the terminal cells express CDX2 and differentiated intestine markers including FABP1 and TFF3.
To annotate the two non-canonical terminal states, we compiled known squamous and neuroendocrine cell markers observed in healthy cells or non-CRC cancers, and calculated the Pearson correlations between their imputed expression and non-intestinal branch probabilities (Extended Data Fig. 8a,b). We excluded all cells with a probability of <0.5 for a given branch when calculating correlations, to avoid interference by cell states outside that branch. This analysis identified branches to neuroendocrine-like and squamous-like states in each patient (a representative example for KG146 is shown in Extended Data Fig. 8a). To further support our annotations, we calculated Pearson correlations between the non-intestinal branch probabilities in each patient and imputed expression for all genes observed in all three patients. We found that the top 5 genes ordered by average correlation among the three squamous branches in KG146, KG182 and KG150 are associated with squamous epithelium and keratinization (DMRTA1, NECTIN4, DLX3, CXCL14, LYPD3), and 4 out of the top 5 genes among the three neuroendocrine branches are associated with glial and neural cells (TRPM3, ITPR2, PLPPR1, PPFIA2) (Supplementary Table 5).
Palantir gene trends were visualized as described in the âVisualization of module trendsâ section using generalized additive models to fit gene expression along Palantir-computed pseudotime (Extended Data Fig. 8c). All expression trends for individual genes were calculated on MAGIC-imputed data (see the âGene denoising and imputationâ section), and the s.d. of each expression bin was represented by the s.d. of the residuals of the fit.
Cell state classifications in KG146 patient tumours
To annotate KG146 tumour cells, we first analysed primary tumour data (880 cells) by determining PCs that explain 75% of the variance (119 PCs) and using PhenoGraph (v.1.5.7) (kâ=â30) to identify six clusters. Repeating this process for the liver metastasis (1,279 cells, 200 PCs) yielded nine clusters. We directly transferred all Hotspot gene module scores from our complete tumour dataset to the KG146 tumour cells, calculated the average Hotspot gene module scores for each cluster and annotated the clusters as ISC-like, absorptive-like, secretory-like, fetal, injury repair, neuroendocrine-like and squamous-like based on high scores for specific gene modules associated with respective labels (Extended Data Fig. 9b,c). This resulted in four ISC-like clusters, one fetal/injury repair cluster and one secretory-like cluster in the primary tumour data, and two ISC-like clusters, one absorptive-like cluster, one secretory-like cluster, two fetal/injury repair clusters, one neuroendocrine cluster and two squamous clusters in the liver metastasis data. In both datasets, we further reclassified one cluster of ISC-like cells as TA/Proliferative-like based on their unique expression of the proliferation markers such as MKI67 and PCNA.
We plotted groups of cells by PhenoGraph (v.1.5.7) cluster (column) and Hotspot module scores (row) using the scanpy dotplot function, with cluster assignments generated within each sample using kâ=â30 (Extended Data Fig. 9d).
Normalizing and scoring gene sets in organoid data
To ensure module and fetal signature scores (Extended Data Figs. 9d and 10a,c,d) were comparable across datasets, we joined our original HISC, IGFF and irinotecan-treated organoid and KG146 patient tumour datasets and normalized the combined data as described in the âData normalization and dimensionality reductionâ section. We then z-scored the log-normalized gene expression matrix across cells and calculated gene set scores on this matrix using the score_genes function in scanpy using lists of genes for each module and the list of fetal signature genes. The shPROX1 organoid datasets were normalized as described in the âData normalization and dimensionality reductionâ section independently of this dataset.
Mapping organoid data to patient tumour
To map cells of each organoid sample to the phenotypically closest tumour cell state from the full KG146 patient dataset, we developed a manifold-based classifier that combines Harmony77, a framework for connecting scRNA-seq data using an affinity matrix augmented by mutual nearest neighbours between datasets, and PhenoGraph11 to transfer labels between datasets. We performed the same analysis on each organoid sample individually along with the relevant matched primary or metastatic patient sample, including original (Fig. 3b), irinotecan-treated (Extended Data Fig. 5b) and shPROX1 (Fig. 4a) samples. The process involves three distinct stepsâfeature selection, co-embedding and classification. The resulting numpy matrices were concatenated to obtain an âaugmentedâ cellâcell affinity matrix that consists of three main components: (1) similarity between in vivo cells; (2) similarity between in vitro cells; (3) similarity between in vitro and in vivo cells.
Feature selection
We identified the top 100 DEGs for each annotated cell state in the KG146 patient tumour dataset, resulting in a total of 800 genes. We then partitioned the resulting lists to retain the subset of highly significant genes with log[FC]â>â3 and BenjaminiâHochberg-adjusted Pâ<â0.001, resulting in 753 genes total. These genes were used for PCA and to produce neighbour graphs in the following steps.
Co-embedding
As expected given the differences between in vivo and in vitro data, using a standard co-embedding approach consisting of a joint PCA and UMAP, we observed extreme batch effects between the two datasets, making label transfer between similar cells ineffective. We therefore followed the approach outlined previously45 to bridge between datasets. We first computed the nearest neighbour graph (Scanpy neighbours function with kâ=â30) in each dataset separately, then computed mutual nearest neighbours (MNNs) between the samples using Harmony77. Importantly, we used the cosine metric to quantify the distance between cells across samples, as this metric is less sensitive to technical artifacts and better reflects conserved biological states in both in vivo and in vitro samples78. We chose a higher number of mutual neighbours (kâ=â60) because it is more robust to sparsity in the MNN graph.
Next, the within-sample nearest neighbour and between-sample MNN graphs were converted to within-sample and between-sample affinity matrices, respectively, using the adaptive Gaussian kernel (default parameters) as implemented in Harmony77. The resulting matrices were concatenated to obtain an augmented cellâcell affinity matrix that consists of three main components: (1) similarity between in vivo cells; (2) similarity between in vitro cells; and (3) similarity between in vitro and in vivo cells. This matrix was input to PhenoGraph (v.1.5.7) classification (see below) to propagate labels from the reference (KG146) dataset to the unlabelled dataset (organoid) and generate UMAP co-embeddings of patient and organoid datasets.
Classification
Finally, we supplied the augmented affinity matrix from step 2 to the PhenoGraph (v.1.5.7) classify function11 with the default parameters. This function converts the affinity matrix into a row-normalized Markov matrix and computes the probability of random walks starting from unlabelled cells from the in vitro samples, and reaching a class of labelled cells in the in vivo sample. Finally, each unlabelled cell is assigned the cell-state label with the maximum probability.
Given the large differences between the in vitro and in vivo samples, we wanted to summarize our classification using coarser cell typing. Thus, to summarize our organoid sample classifications, we aggregated the probabilities of different cell states into three generalized categories; ISC-like, combining TA/Proliferative for ISC/TA, absorptive-like and secretory-like for differentiated intestine, and combining fetal/injury repair, neuroendocrine and squamous for non-canonical. These groupings can be interpreted as the likelihood of a cell belonging to any of the several cell states which were combined. The probabilities for each resulting categories are plotted using the python-ternary (v.1.0.8)79 package (Fig. 3b and Extended Data Fig. 10b).
Identification of fetal-state-associated transcription factors
We aimed to generate a ranked list of human transcription factors that are associated with the fetal progenitor state in a conserved manner across non-canonical patient tumours. Starting with a list of 1,665 human transcription factors80, we restricted potential targets to transcription factors for which we observe (1) greater than 10 UMIs total in all four patient datasets (leaving 1,099 transcription factors); (2) at least 5 UMIs in any cell in all four patient datasets (leaving 527 transcription factors); and (3) at least 50 transcription factor-expressing cells in all four patient datasets (leaving 508 transcription factors). This filtering was intended to remove sparsely expressed transcription factors that may not be as easily targeted, and to restrict our analysis to transcription factors that are more reliably correlated with our fetal signature.
Next, we used Palantir to compute expression trends for all the transcription factors and the fetal progenitor signature score along the canonical-to-non-canonical DC (see the âDelineation of canonical to non-canonical tumour axes across patientsâ section). As we are interested in transcription factors that drive the transition from canonical to fetal cell states, we focused on those with peak expression just prior to entering non-canonical states along the DC of each patient. For patients KG146, KG182, KG150 and KG183, we first identified the position along their DC of the first maxima of the trend in the fetal progenitor signature calculated along this DC (indicated by the vertical dashed lines in Extended Data Fig. 7a). Maxima were identified as the first inflection point along the first derivative of the trend (that is, when the derivative first changes from positive to negative). Trends for KG150 and KG183 lack a first-derivative inflection point, so we used the position of the maximum value for these patients. We then calculated the Pearson correlation between the expression of each transcription factor and the fetal progenitor gene signature score using only cells at positions along a patientâs DC which precede the signature score peak. This yielded four correlation values total for each transcription factor, one for each patient. We focused only on transcription factors with a minimum correlation of râ=â0.5 in patient KG146 (leaving 14 transcription factors) and râ=â0.2 in all four patients (leaving 5 transcription factors).
For the remaining six transcription factors, we determined their treatment response in HISC-grown organoids by computing the log-transformed fold change between irinotecan-treated and untreated conditions. log-transformed fold changes were calculated from single-cell data only using cells classified as non-canonical (see the âMapping organoid data to patient tumourâ section) (Extended Data Fig. 11b).
Multiplexed immunofluorescence
Multiplexed tissue staining and imaging
To maximize capture of all areas of the tumour including the invasion front, full clinical pathology sections prepared for clinical diagnosis were used for imaging (not core punches as typically used for tissue microarrays). Primary antibody staining conditions were optimized using standard immunohistochemical staining on the Leica Bond RX automated research stainer with DAB detection (Leica Bond Polymer Refine Detection, DS9800). Using 4âµm FFPE tissue sections and serial antibody titrations, the optimal antibody concentration was determined followed by transition to a seven-colour multiplex assay with equivalency. Optimal primary antibody stripping conditions between rounds in the seven-colour assay were performed following one cycle of tyramide deposition followed by heat-induced stripping (see below) and subsequent chromogenic development (Leica Bond Polymer Regine Detection, DS9800) with visual inspection for chromogenic product with a light microscope (T.J.H.). Multiplex assay antibodies and conditions are described in Supplementary Table 8.
Seven-colour imaging assay
FFPE 4âµm tissue sections of were baked for 2.5âh at 63â°C in vertical slide orientation with subsequent deparaffinization performed on the Leica Bond RX followed by 30âmin of antigen retrieval with Leica Bond ER2 followed by six sequential cycles of staining with each round including a 30âmin combined block and primary antibody incubation (Akoya antibody diluent/block ARD1001EA), except for HER2, which required a 1âh incubation. From each tissue section, we captured around nine fields of view (FOVs) on average, each 1.34âmm2 in size.
For chromogranin A and OLFM4, detection was performed using a secondary horseradish peroxidase (HRP)-conjugated polymer (Akoya Opal polymer HRP Ms + Rb ARH1001EA; 1:5, 10âmin incubation). Detection of all of the other primary antibodies was performed using goat anti-rabbit Poly HRP secondary antibody (Invitrogen, B40962, 1:100, 10âmin incubation). The HRP-conjugated secondary antibody polymer was detected using fluorescent tyramide signal amplification using Opal dyes 520, 540, 570, 620, 650 and 690 (Akoya, FP1487001KT, FP1494001KT, FP1488001KT, FP1495001KT, FP1496001KT, FP1497001KT). The covalent tyramide reaction was followed by heat-induced stripping of the primaryâsecondary antibody complex using Akoya AR9 buffer (AR900250ML) and Leica Bond ER2 (90% AR9 and 10% ER2) at 100â°C for 20âmin preceding the next cycle (1 cycle of stripping for HER2 (1âμgâmlâ1, CST, D8F12), CDX2 (0.46âμgâmlâ1, CST, D11D10), TP63 (0.15âμgâmlâ1, CST, D9L7L), PLCG2 (0.2âμgâmlâ1, CST, E5U4T) and chromogranin A (2.705âμgâmlâ1, Abcam, EP1030Y); 2 cycles for SOX2 (1.26âμgâmlâ1, Abcam, SP76), CK20 (0.208âμgâmlâ1, CST, D9Z1Z), VIM (0.0375âμgâmlâ1, CST, D21H3), TROP2 (3.29âμgâmlâ1, Abcam, SP294), CK5 (0.142âμgâmlâ1, CST, E2T4B) and OLFM4 (0.27âμgâmlâ1, CST, D1E4M); 2.5 cycles of stripping for Ki-67 (1:100, Biocare, SP6). After six sequential rounds of staining, the sections were stained with Hoechst (Invitrogen, 33342) to visualize nuclei and mounted with ProLong Gold antifade reagent mounting medium (Invitrogen, P36930).
Multispectral imaging and spectral unmixing
Seven-colour multiplex-stained slides were imaged using the Vectra Multispectral Imaging System version 3 (Akoya). Scanning was performed at Ã20 (Ã200 final magnification). Filter cubes used for multispectral imaging were DAPI, FITC, Cy3, Texas Red and Cy5. A spectral library containing the emitted spectral peaks of the fluorophores in this study was created using the Vectra image analysis software (Akoya). Using multispectral images from single-stained slides for each marker, the spectral library was used to separate each multispectral cube into individual components (spectral unmixing) allowing for identification of the seven marker channels of interest using Inform v.2.4 image analysis software.
Twelve-colour imaging assay
FFPE 4âµm tissue sections of were baked for 1âh at 62â°C in vertical slide orientation with subsequent prestaining of deparaffinization (Leica, Bond Dewax Solution, AR9222) and 35âmin of Bond ER2 (Leica, AR9640) antigen retrieval at 100â°C was performed on the Leica Bond RX automated research strainer. The Lunaphore COMET imaging chip (MK03) was placed over the acquisition region based on alignment with regions selected on a haematoxylin and eosin stained image, then loaded into the COMET device and subjected to a first cycle of autofluorescence image acquisition, followed by nine cycles of staining, imaging and elution. Each cycle contained one rabbit primary antibody and/or one mouse primary antibody (HER2 (1âμgâmlâ1, CST, D8F12); CDX2 (0.46âμgâmlâ1, CST, D11D10); PROX1 (1.5âμgâmlâ1, CST, D2J6J); Ki-67 (1âμgâmlâ1, Abcam, EPR3610); vimentin (0.33âμgâmlâ1, Thermo Fisher Scientific, V9); PLCG2 (1âμgâmlâ1, CST, E5U4T); PLCG2 (1âμgâmlâ1, CST, E5U4T); CHGA (0.13âμgâmlâ1, CST, 5H7); CK5 (0.213âμgâmlâ1, CST, E2T4B); SYNC (2âμgâmlâ1, Thermo Fisher Scientific, Poly); CK20 (0.21âμgâmlâ1, CST, D9Z1Z); GPC1 (6.47âμgâmlâ1, Abcam, EPR22580-72)) detected with an Alexa Flour conjugated species-specific secondary antibody (Invitrogen, 10âµgâmlâ1; Supplementary Table 8). The cyclic staining, imaging and elution was automatically performed by COMET. The optimal concentration, position in the panel and incubation time (4âmin for each marker per cycle, but 8âmin for HER2) of each marker in the panel was optimized to be concordant with its corresponding single diaminobenzidine immunohistochemistry stain. Optimal immunohistochemistry staining for each marker was determined by an MSK pathologist (T.H.) according to the DAB staining result, as performed on the Leica BondRX Stainer using the BOND Polymer Refine Detection Kit (Leica, DS9800). All antibodies were pre-diluted for COMET analysis with Intercept Antibody Diluent (LI-COR, 927-65001). After each staining cycle, elution of primary and secondary antibodies was performed using elution buffer solution (Lunaphore, BU07-L). For each sample, a 82.5âmm2 region of interest was imaged under the Lunaphore slide cover chip at Ã20 magnification, and a full channel stacked OME.tif file was generated automatically once all cycles were completed.
Single-cell segmentation
We used Mesmer (v.0.12)81, a deep-learning cell segmentation algorithm, to identify cell boundaries in all COMET and Vectra images. The input to Mesmer (v.0.12) is a single nucleus-stained image and a single membrane or cytoplasm-stained image to define the extent of each nucleus and cell. We used DAPI as a nuclear marker for all COMET and Vectra images. To create an image that will define the boundaries of multiple cell types, we combined the channels for several cell-type-specific membrane or cytoplasmic markers into a single image by minâmax scaling each channel (using the MinMaxScaler function in the sklearn.preprocessing (v.1.4.2) package with the default parameters) and summing them. For COMET, we combined CK20, HER2, CK5, SYNC (normal and tumour epithelial cells) and VIM (stromal cells). For Vectra panel 1, we used HER2, SOX2, CK20, CDX2 and CHGA (tumour cells) and VIM (stromal cells). For Vectra panel 2, we used TP63, OLFM4, TROP2 and CK5 (tumour cells) and PLCG2 (stromal cells).
We ran Mesmer (v.0.12) on these images with the default parameters to predict cell boundaries, and calculated the cell size, eccentricity and centroid of each cell boundary using the regionprops function (default parameters) in the Python skimage (v.0.23.2) package. When running Mesmer (v.0.12), we first subsampled COMET images by half so that they would fit in system memory (128âGB of memory). For both cell size and DAPI expression, we found the distributions of segmented cells produced by Mesmer (v.0.12) were bimodal and the lower mode contained primarily empty regions and not real cells. We therefore filtered out all predicted cell boundaries below threshold values of 30âpx2 cell size (estimated from the distribution) and a log2-normalized DAPI intensity of 11 (COMET), 1 (Vectra panel 1) and 1 (Vectra panel 2) (estimated from the distribution). This resulted in a COMET dataset of 6,852,690 cells across 18 FOVs, a Vectra panel 1 dataset of 6,090,968 cells across 664 FOVs; and a Vectra panel 2 dataset of 5,213,051 cells across 602 FOVs.
Normalization, background removal and thresholding
Raw per-cell marker expression levels were first determined by summing the brightness values over all pixels within each cell boundary. To ensure that our downstream analysis was not influenced by cell size, we then divided the per-cell expression for each channel by the cell boundary sizes determined by the regionprops function, as described above. Once normalized, all cells were pooled into cell-by-expression matrices within the same imaging technology and panel (Vectra panel 1, Vectra panel 2 and COMET) for downstream analyses and annotated with patient- and sample-level metadata.
CK5 background in Vectra panel 2
To address the high levels of background in CK5 staining, an experienced physician manually annotated 167 FOVs out of 602 to denote whether they have high background signal. To identify high-background images in the remaining 435 FOVs, we first found the highest level of background CK5 signal (10th percentile of expression across all cells in the FOV) within FOVs labelled low-background (around 0.0068). We then classified all unlabelled FOVs with a background CK5 signal of >0.0068 as high background. As a result, 327 unlabelled FOVs with low background were used in later analyses, and 108 with high background were removed. In total, we used 456 FOVs with a low level of CK5 background expression in all figures involving Vectra panel 2.
Thresholding for tumour markers in Vectra panels 1 and 2
To call cells as âpositiveâ for the tumour markers in each panel, we calculated the quantile values of expression across all cells in small increments from 0.0 to 1.0 and found the knee-point of these values (typically between 0.8 and 0.9 for all markers). This method enabled us to avoid using a single quantile threshold for all markers, which would be unreasonable, as markers have different expression distributions across samples. The intuition behind our approach is that the knee-point at which quantile values start increasing rapidly reflects where the population changes from negative to positive. This can also be seen as a way of finding the knee-point in the distribution without fitting an estimated distribution to the data. We validated our labels by comparing the expression of tumour markers with tumour regions that were manually annotated by a physician in a subset of images. We then took all cells called as positive for a marker and repeated the knee-point analysis to identify the subset of cells with high marker expression, which we report as a percentage of all cells positive for any tumour marker, listed above in the âSingle-cell segmentationâ section, from the same panel; we quantify CK20 in panel 1 and OLFM4, TROP2 and CK5 in panel 2 this manner (Extended Data Figs. 3f and 5aâd).
PROX1+ and CDX2+ cell identification in COMET
For each COMET image, regions corresponding to normal epithelium and tumour tissue were manually demarcated by an experienced physician based on histology, and we retained only cells belonging to these masked regions for downstream analyses. We included this preliminary filtering step to (1) remove most stromal cells, therefore reducing the computational cost downstream; and (2) distinguish tumour cells from normal epithelium using histological evidence, as this was not possible from marker expression alone (for example, due to expression of stromal marker genes in basal-like tumour cells). As we use only COMET data to compare CDX2+ intestinal and PROX1+ fetal-like cells, we first restricted our analysis to cells expressing either PROX1 or CDX2 above a stringent level of background (99th percentile of expression in unmasked regions calculated separately for each image). We then removed 6 samples with fewer than 1% PROX1-expressing cells, leaving 7 samples. For each of these samples, we further labelled cells as CDX2+, PROX1+ or double+ if their expression of one or both genes was >0.5âs.d. above the mean.
Organoid experimental methods
Organoid and cell culture
For experiments, organoids were collected from Matrigel with 3âmM EDTA in DPBS and, where indicated, were treated with TrypLE (Thermo Fisher Scientific) for 5â10âmin at 37â°C and filtered through a 40-μm cell strainer to generate single cells. Cells were plated at a density of 2,000 cells per 40âμl of Matrigel and were incubated at 37â°C for 30âmin until the domes solidified. Organoids were cultured in HISC (Advanced DMEM/F12 (AdDF12; Thermo Fisher Scientific), GlutaMAX (2âmM, Thermo Fisher Scientific), HEPES (10âmM, Thermo Fisher Scientific), N-acetyl-l-cysteine (1âmM, Sigma-Aldrich), B27 supplement with vitamin A (Thermo Fisher Scientific), primocin (100âμgâmlâ1, InvivoGen), EGF (50ângâmlâ1, Peprotech), Noggin (100ângâmlâ1, Peprotech), A8301 (500ânM, Sigma-Aldrich), FGF2 (50ângâmlâ1, Peprotech), IGF-I (100ângâmlâ1, Peprotech)) or IGFF (HISC without EGF, Noggin, A-8301, FGF2, IGF-I) medium supplemented with Y-27632. The medium was changed every 3â4âdays. Organoids were collected at 7âdays for downstream assays. Where indicated, small intact organoids (2â3âdays after passage) were treated with 250ânM irinotecan added to HISC medium for 7âdays and then collected for downstream assays. Lentiviral production and transduction of organoids was performed as described previously82. In brief, HEK293T cells (ATCC) were cultured in DMEM supplemented with 10% FBS, GlutaMAX (2âmM, Thermo Fisher Scientific) and penicillinâstreptomycin (100âIUâmlâ1, 0.1âmgâmlâ1, Thermo Fisher Scientific). All cells tested negative for mycoplasma. For scRNA-seq, organoids were collected from Matrigel with 3âmM EDTA in DPBS, dissociated to single cells with Accutase (Sigma-Aldrich) for 30â45âmin at room temperature, washed with IGFF medium and processed as described above.
Inducible knockdown
For doxycycline inducible PROX1 knockdown experiments, de novo 97-mer mirE shRNA sequences were synthesized (IDT Ultramers) and PCR amplified using the primers miRE-Xho-fw (5â²-TGAACTCGAGAAGGTATATTGCTGTTGACAGTGAGCG-3â²) and miRE-EcoOligo-rev (5â²-TCTCGAATTCTAGCCCCTTGAAGTCCGAGGCAGTAGGC-3â²) as described previously83. Sequences (shPROX1-1: TGCTGTTGACAGTGAGCGCGAGGACCAAGATGTCATCTCATAGTGAAGCCACAGATGTATGAGATGACATCTTGGTCCTCATGCCTACTGCCTCGGA; shPROX1-2: TGCTGTTGACAGTGAGCGCCCCCGAGAAAGTTACAGAGAATAGTGAAGCCACAGATGTATTCTCTGTAACTTTCTCGGGGATGCCTACTGCCTCGGA) were cloned into the LT3GEPIR backbone83 (Addgene, 111177) and used to generate lentiviral particles to transduce into organoids as described previously82. The original plasmid containing an shRNA sequence to Renilla 306 luciferase (shRen.713) was used as a control. Transduced organoids were selected using HISC medium supplemented with 2âμgâmlâ1 puromycin for 7âdays. For inducible knockdown experiments, organoids were dissociated into single cells, plated at a density of 2,000 cells per 40âμl of Matrigel and maintained in HISC or IGFF medium supplemented with 2âμgâmlâ1 doxycycline (Thermo Fisher Scientific) unless otherwise specified for 7âdays before downstream assays. For organoid initiation and outgrowth assays, organoids containing inducible PROX1 or control shRNA cultured in HISC medium supplemented with 2âμgâmlâ1 doxycycline for 7âdays were dissociated into single cells, stained with DAPI (1âμgâmlâ1, Thermo Fisher Scientific), and live cell (DAPIâ) and GFP+ sorted to select for healthy cells expressing the shRNA construct, and plated at a density of 750 cells per 15âμl Matrigel in HISC medium without Y-27632 and supplemented with 2âμgâmlâ1 doxycycline, and imaged at 7âdays (BioTek).
Western blotting
Approximately 4,000 organoids (3â4âmillion cells) were recovered from Matrigel using 3âmM EDTA in DPBS, washed, centrifuged (200g, 5âmin, 4â°C) and lysed with 1à RIPA buffer supplemented with PPI (1:100, Sigma-Aldrich, 04693132001) and benzonase (1:100, Thermo Fisher Scientific, 70-664-3) on ice for 30âmin. The protein concentration was determined using Pierce BCA assay (Thermo Fisher Scientific, 23227). A total of 10âμg protein per sample was separated by SDSâPAGE on Bis-Tris polyacrylamide gels (Thermo Fisher Scientific, NW04120BOX), transferred to activated PVDF membranes (Millipore, IPFL00010) and blocked in 3% BSA-TBST solution for 30âmin. The membranes were incubated overnight at 4â°C with the following antibodies: mouse anti-β-actin (1:1,000, Thermo Fisher Scientific, AM4302) and rabbit anti-PROX1 (1:1,000, Abcam, ab199359), followed by secondary antibody incubation with 488 anti-mouse and 680 anti-rabbit secondary antibodies (1:5,000, LI-COR Biosciences, 1âh, room temperature) before imaging (Odyssey CLx). Western blots were quantified using ImageJ (v.1.53t)84.
Organoid whole-mount immunofluorescence
Whole organoids were plated in 10% Matrigel and HISC medium on chamber slides with removable wells (Thermo Fisher Scientific, 177380), and incubated at 37â°C for 1âh. The organoids were then fixed in 4% paraformaldehyde in PME solution for 10âmin, washed twice with IF buffer (0.2% Triton X-100, 0.05% Tween-20 in DPBS), permeabilized (0.5% Triton X-100 in DPBS for 10âmin, room temperature), blocked in 10% normal goat serum (Thermo Fisher Scientific, 50062Z) for 30âmin before overnight primary antibody incubation with 1:500 rabbit anti-PROX1 (Abcam, ab199359) in 10% normal goat serum at 4â°C. Organoids were incubated with 1:400 of 594 anti-rabbit antibody (Invitrogen, A11012) for 1âh at room temperature, washed three times with IF buffer and one time with cold DPBS. The wells were removed and the samples mounted using medium containing DAPI (Novus Biologics, H-1200-NB). Imaging was performed on the Zeiss Axio Imager 2 with Apotome structured illumination microscope. PROX1 expression was quantified using the CellProfiler (v.4.2.5) software85.
RTâqPCR
Total RNA was extracted from organoids using the RNeasy Mini kit (Qiagen). Total RNA (3â4âμg) was used to prepare cDNA using the Transcriptor First-Strand cDNA synthesis kit (Roche). qPCR was performed with TaqMan gene expression assay primers (Thermo Fisher Scientific; PROX1, Hs00896293_m1; GAPDH, Hs02758991_g1; ELF5, Hs01063022_m1; KRT23, Hs00210096_m1; TFF3, Hs00902278_m1; FABP1, Hs00155026_m1; TRPS1, Hs00936363_m1; GNAI1, Hs01053355_m1; FGF20, Hs00173929_m1; NELL2, Hs00196254_m1; NRXN3, Hs01028186_m1; POMC, Hs01596743_m1; KRT20, Hs00300643_m1; GPC1, Hs00892476_m1; LGR5, Hs00173664_m1; TMEM132A, Hs01096434_m1; NEUROD1, Hs01922995_s1; CHGB, Hs01084631_m1), the Relative expression was quantified using the ÎÎCt method, normalized to the expression of GAPDH on the QuantStudio 6 and 7 Pro real-time PCR system (Applied Biosystems).
Orthotopic xenograft experiments
All animal experiments were performed in accordance with protocols approved by the Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committee (IACUC). NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ, 005557) mice were obtained from the Jackson Laboratory and were transplanted at 6 weeks of age86. Mice were maintained in a specific-pathogen-free (SPF) facility under a 12âhâ12âh lightâdark cycle under controlled temperature and humidity, and given ad libitum access to standard diet or irradiated diet supplemented with 2,500âppm doxycycline (Modified LabDiet 5053) and water. For orthotopic caecal and intrahepatic injections, OKG146P, OKG146Li, OKG136P or OKG136Li organoids were transduced with pLenti-PGK-Akaluc (AkaLuc) or pLenti-PGK-tdTomato-Akaluc (TdT-AkaLuc) virus (subcloned from pLenti-PGK-Venus-Akaluc(neo)) and HR180-LGR5-iCT plasmids through Gibson assembly (Addgene, 124701, 129094). For intrahepatic injections assessing PROX1 knockdown, OKG146P and OKG136P lines expressing shRNAs to PROX1 or Renilla control were transduced with pLenti-PGK-Akaluc (AkaLuc). For caecal injections, 200,000 cells from each organoid line were mixed with 50% Matrigel in 10âμl of HISC and injected into the caecal submucosa of NSG mice. For hepatic injections, 500,000 cells from each organoid line were mixed with 50% Matrigel in 10âμl of HISC and injected under the liver capsule of NSG mice. Weekly bioluminescence imaging was performed on the IVIS Spectrum Xenogen instrument (Caliper Life Sciences) and analysed using Living Image software v.2.50. Experimental group sizes were practically determined on the basis of five mice per cage, with nâââ¥ââ5 age- and sex-matched mice per group. Animals were randomly assigned to experimental groups where relevant. Experimental end point was reached when tumour size exceeded >10% of body mass, or when animals showed any signs of respiratory distress or illness, such as hunched posture, failure to groom or greater than 10â15% weight loss. These limits were not exceeded in any of the experiments. The animals were euthanized at the humane end point, and tissues were collected for downstream assays. Where needed, tissues were fixed in 4% paraformaldehyde. Investigators were blinded to mouse group when scoring histopathological samples.
Statistics and reproducibility
In vitro experiments were repeated a minimum of three times independently with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.