Cell lines
The B16F10 melanoma (ATCC CRL-6475), CT26 colon carcinoma (ATCC CRL-2638) and 4T1 breast cancer (ATCC CRL-2539) authenticated cell lines were purchased directly from ATCC. CT26-Luc, B16F10-Luc and 4T1-Luc cells were lentivirally transduced with luciferase. Cells were confirmed mycoplasma free. Cells were cultured in incubators at 37â°C with atmosphere of humidified 5% CO2. B16F10 and B16F10-Luc cells were grown in DMEM supplemented with 10% (vol/vol) fetal bovine serum (FBS), 1à GlutaMax, 1% (vol/vol) MEM non-essential amino acids solution (Gibco-11140050) and 100âUâmlâ1 penicillinâstreptomycin. CT26, CT26-Luc, 4T1 and 4T1-Luc cells were grown in Roswell Park Memorial Institute medium (RPMI-1640) supplemented with 10% (vol/vol) FBS, 1à GlutaMax, 1% (vol/vol) MEM non-essential amino acids solution and 100âUâmlâ1 penicillinâstreptomycin. No commonly misidentified cell lines were used in this study.
Exome sequencing
Paired tumour and tail DNA from BALB/c mice bearing subcutaneous CT26 tumours or C57BL/6 mice bearing subcutaneous B16F10 tumours was extracted in triplicate (nâ=â3 mice per tumour line) using Qiagen DNeasy Blood & Tissue Minikit following the manufacturerâs instructions. Exome capture from mouse tumour and tail DNA triplicates was conducted using Agilent SureSelectXT All Exon kit for target enrichment DNA library preparation56, according to the manufacturerâs instructions (Agilent). Genomic DNA was fragmented by acoustic shearing with a Covaris S220 instrument. Fragmented DNAs were cleaned, end-repaired and adenylated at the 3â² end. Adaptors were ligated to DNA fragments, and adaptor-ligated DNA fragments enriched with limited-cycle PCR. Adaptor-ligated DNA fragments were validated using Agilent TapeStation (Agilent) and quantified using Qubit 2.0 Fluorometer (ThermoFisher Scientific) and Real-Time PCR (KAPA Biosystems). Sequencing libraries were clustered onto a lane of a flow cell. After clustering, the flow cell was loaded on an Illumina HiSeq4000 Instrument per the manufacturerâs instructions. Samples were sequenced using 2âÃâ150âbp paired end configuration. Image analysis and base calling was conducted by the HiSeq Control Software. Raw sequence data (.bcl files) generated from Illumina HiSeq was converted into fastq files and de-multiplexed using Illumina bcl2fastq2.17. Sequence reads were trimmed to remove adaptor sequences and nucleotides with poor quality using Trimmomatic v.0.39 (ref. 57). Trimmed reads were aligned to the GRCm38 reference genome using the Illumina Dragen Bio-IT platform. Alignments were sorted and PCR or optical duplicates marked for generation of BAM files. Somatic single-nucleotide variants and insertion or deletion (indel) variants were called using Illumina Dragen58 and GATK Mutect2 (ref. 59). All variants from paired-normal tissue and murine variants from the dbSNP database60 were removed during the process. VCF files were left aligned and normalized, with splitting of multiallelic sites into several sites using bcftools v.1.13 (ref. 61). Only tumour-specific variants called by both algorithms were used for further analysis.
RNA sequencing
Tumour RNA from BALB/c mice bearing subcutaneous CT26 tumours or C57BL/6 mice bearing subcutaneous B16F10 tumours was extracted in triplicate using Qiagen RNeasy Minikit as per the manufacturerâs instructions. Extracted RNA samples were quantified using Qubit 2.0 Fluorometer (Life Technologies) and RNA integrity checked using Agilent TapeStation 2400 (Agilent). RNA sequencing libraries were prepared using the NEBNext Ultra RNA library Prep Kit for Illumina as per the manufacturerâs instructions (New England Biolabs). mRNAs were enriched with Oligo(dT) beads. Enriched mRNAs were fragmented for 15âmin at 94â°C. First- and second-strand complementary DNAs (cDNAs) were synthesized subsequently. cDNA fragments were end-repaired and adenylated at 3â² ends, and universal adaptors ligated to cDNA fragments, followed by index addition and library enrichment by limited-cycle PCR. Sequencing libraries were validated on Agilent TapeStation (Agilent), and quantified using Qubit 2.0 Fluorometer (Invitrogen) and quantitative PCR (qPCR) (KAPA Biosystems). Library loading, sequencing and read trimming were done as described above. Trimmed reads were aligned to the mm10 reference using STAR aligner v.2.5.2b (ref. 62). Unique gene hit counts were calculated using feature counts from Subread Package v.1.5.2. Unique reads that fell within exon regions were counted. The gene hit counts table was used for expression analysis using DESeq2 v.1.20.0 (ref. 63).
Neoantigen prediction and selection
Mutation-specific RNA expression and allele fraction were added to somatic VCF files using Bam-readcount64 and VAtools (http://vatools.org). Somatic VCFs were annotated with The Ensembl Variant Effect Predictor (VEP Ensembl v.104)65. Only PASS variants from VCFs were considered. Annotated VCFs were analysed using pVacSeq for neoepitope discovery14. MHC-I affinities were predicted with NetMHCpan v.4.1 (ref. 66) and NetMHC v.4.0 (ref. 67), and MHC-II affinities were predicted with NetMHCIIpan v.4.1 (ref. 68) and NNalign v.2.0 (ref. 69). Exonic mutation-derived long peptides based on single-nucleotide polymorphisms (SNP) or indels predicted to generate mutant MHC-binding peptides were first filtered on the basis of the set of minimum criteria: (1) present in all tumour sample triplicates (DNA variant allele fractionââ¥â0.05) and none of the normal tissue triplicates, (2) non-synonymous mutation resulting from either SNP or indel, (3) confirmed exonic mutation transcription (RNA variant allele fractionââ¥â0.05) and gene expression by RNA sequencing in tumour sample triplicate (transcripts per millionââ¥â1), (4) at least one predicted MHC-I or MHC-II binding epitope and (5) MHC-I or MHC-II differential binding affinity17,20 (wild-type half-maximum inhibitory concentration (IC50)/mutant IC50)ââ¥â1.2. Predicted neoantigens fulfilling all previous criteria were then prioritized for inclusion and selected according to the following hierarchy: (1) high predicted affinity (MHC-I or MHC-II IC50ââ¤â500ânM), (2) moderate predicted affinity (MHC-I or MHC-II IC50 500â1,000ânM) and (3) low predicted affinity (MHC-I or MHC-II IC50 1,000â5,000ânM) (Extended Data Tables 1 and 2).
Strains and plasmids
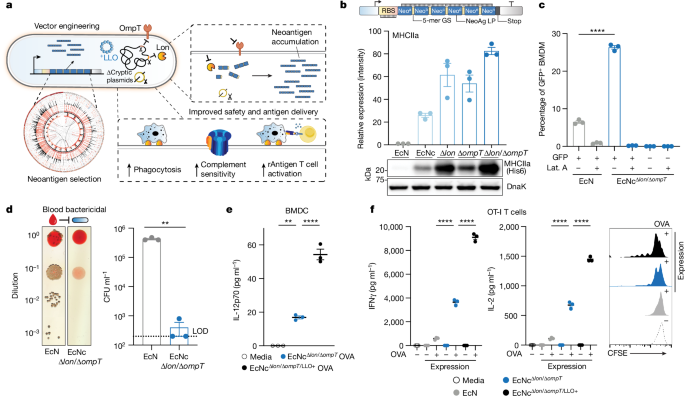
Plasmids were constructed using restriction-enzyme mediated and Gibson assembly cloning methods. Neoantigen construct iterations were designed and created as Geneblocks (IDT) encoding a constitutive promoter and 5â² untranslated region (UTR) containing selected ribosome-binding site, followed by coding region composed of mutant-residue containing long peptides connected in tandem or by various linkers as indicated. 5â² BamHI and 3â² XbaI restriction endonuclease sites were added to constructs. Coding sequences were codon optimized for E. coli. Constructs were cloned between BamHI and XbaI restriction sites on a stabilized p246-luxCDABE plasmid where luxCDABE had been cloned out22, and flanked by 3Ⲡλ-phage transcription terminator, with high-copy pUC origin. For protein expression assessment studies, the codon sequence for a 6Ã-Histidine Tag (HisTag) was added immediately before the stop codon within the neoantigen construct coding sequence by PCR amplification of full construct plasmids with oligonucleotide containing 6Ã-HisTag sequence followed by kinase, ligase, DpnI enzyme mix protocol (NEB). Neoantigen construct plasmids were transformed into chemically competent E. coli DH5α or BL21(DE3) (New England Biolabs), or electrocompetent EcN parental strain or genetic derivatives. The parental EcN strain and all derivatives used in this study harbour an integrated luxCDABE cassette within the genome, which also contains an erythromycin resistance gene70. Plasmid encoding constitutive LLO was constructed by cloning in the hok/sok stabilization system to pCG02-p15a backbone71, PCR amplification of backbone with SLC cloned out, and Gibson assembly of Geneblock encoding LLO under constitutive promoter and 5â² UTR containing selected ribosome-binding site. Constitutive LLO plasmids were transformed into electrocompetent EcN parental and genetic derivative strains. Strains were cultured in Luria-Bertani (LB) medium with antibiotics for plasmid retention (pUC:kanamycin 50âμgâmlâ1, p15a:spectinomycin 50âμgâmlâ1) in a 37â°C orbital incubator.
Construction of cryptic plasmid-cured EcN
EcN cryptic plasmids were cured with Cas9-mediated double-strand break, as described previously27. Briefly, EcN was transformed with pFREE or pCryptDel4.8 to cure the cryptic plasmids pMUT1 or pMUT2, respectively. The transformants were grown overnight and diluted 1:1,000 the next day into fresh LB containing 0.2% rhamnose and 0.43âμM anhydrotetracycline. After 24âh of incubation, the culture was streaked onto LB plates without antibiotics and incubated overnight in a 30â°C incubator. Colonies were screened with colony PCR to verify the loss of cryptic plasmids.
Construction of genetic knockout strains
Genetic knockouts were performed using the lambda red recombination system72. In brief, EcNc was transformed with pKD46. Transformants were grown at 30â°C in LB with ampicillin and l-arabinose, then made electrocompetent. The chloramphenicol resistance cassette with corresponding overhangs for each target gene for deletion was prepared by PCR amplification of pKD3. Electroporation was performed using 100âμl of competent cells and 50â300âng amplified DNA. After 2âh of recovery, cells were plated on LB agar containing chloramphenicol and incubated at 37â°C overnight. Target gene deletion was verified by colony PCR. For excision of the antibiotic resistance marker, pCP20 was transformed, and the transformants were plated on fresh LB plates containing ampicillin and incubated at 30â°C overnight. Selected colonies were then inoculated onto fresh LB plates without antibiotics and cultured at 43â°C overnight for induction of flippase and plasmid curing. Clones were subsequently screened for loss of antibiotic resistance.
qPCR for PCN
Copy number variant plasmids were constructed from a high-copy pUC-GFP22 plasmid. The plasmid backbone excluding the pUC origin was PCR-amplified and Gibson assembled with sc101*, p15A or ColE1 origin of replication insert. The respective inserts were prepared from PCR amplification of template plasmid pCG02_sc101*, pCG02_p15A or pTH05_ColE1. Plasmid copy number (PCN) was determined as reported previously22, in which the relative abundance of plasmid DNA compared to genomic DNA is measured by qPCR. Briefly, strains with the plasmid of interest were grown at 37â°C overnight in fresh LB with appropriate antibiotics. Cells were collected by centrifugation at 3,000g at 4â°C for 10âmin, the supernatant removed and the cell pellet resuspended in distilled water for optical density measurement at 600ânm (OD600) equal to 1. Resuspended cells were fivefold serially diluted. Samples were denatured at 95â°C for 10âmin and 2âμl of each sample dilution was added into 18âμl of NEB Luna Universal qPCR Master Mix in each well of a 96-well plate. Then 25-fold diluted samples were used for the measurement of crossing point values: the cycle number when amplified sample fluorescence exceeds the background. Fivefold diluted samples were used for generation of the standard curve for PCR efficiency (E). E was defined from the slope (S) of each standard curve with the equation Eâ=â5(â1/S) and PCN was determined with the equation PCNâ=â(EGCTG)/(EPCTP), where respective values for genomic DNA are denoted by a subscript G and plasmid DNA by subscript P.
Immunoblot and ELISA
For immunoblot and ELISA, a C-terminal 6Ã-HisTag was attached to each neoantigen construct. Strains expressing neoantigen construct with C-terminal 6Ã-HisTag were grown overnight in LB media with appropriate antibiotics. Equalization of OD600 measurement to match colony-forming units (CFU) per ml (CFUâmlâ1) between all cultures was done before all sample processing. CFU-matched cultures were centrifuged at 3,000g at 4â°C for 10âmin. For immunoblot, samples were resuspended in B-PER lysis reagent (ThermoFisher Scientific) containing 250âUâmlâ1 benzonase nuclease (Millipore Sigma) and 1âUâmlâ1 rLysozyme (Millipore Sigma) and placed on an orbital shaker for 15âmin at room temperature. Samples were centrifuged at 10,000g for 20âmin at 4â°C to separate soluble and insoluble fractions or total lysate used directly. Processed samples were mixed with SDS-loading buffer with 5âmM dithiothreitol, boiled and subject to immunoblot analysis. For relative quantification of immunoblot chemiluminescent intensity, target protein bands on the same blot were normalized to the loading control band DnaK for the same sample. DnaK loading controls were always run on the same gel as target proteins. Normalized values were divided to provide relative intensity values. Mouse anti-6ÃHis (αTHE) was purchased from GenScript, mouse anti-DnaK was purchased from Abcam (8E2/2). αTHE and 8E2/2 antibodies were used at 1:5,000 dilution.
For HisTag ELISA, samples were resuspended in ice-cold PBS containing HALT protease inhibitor cocktail (ThermoFisher Scientific). Samples were sonicated on ice for 2âmin total time. Sonicated samples were centrifuged at 10,000g for 20âmin at 4â°C. Soluble sample fractions were analysed using GenScript HisTag ELISA Detection Kit as per the manufacturerâs instructions.
For ex vivo immunoblot analysis, BALB/c mice bearing established hind-flank CT26 tumours were injected intravenously with the EcNcÎlon/ÎompT/LLO+ nAg19-His strain cocktail that contains all three neoantigen constructs, in which each construct (MHCIa, MHCIIa and MHCI/IIv) contained a C-terminal 6Ã-HisTag. Then 48âh after treatment, tumours and TDLNs were extracted from mice and placed in B-PER lysis reagent (ThermoFisher Scientific) with 250âUâmlâ1 benzonase nuclease (Millipore Sigma), and homogenized using a gentleMACS tissue dissociator (Miltenyi Biotec, C-tubes). Tissue homogenate was sonicated on ice for 3âmin. Sonicated samples were centrifuged at 10,000g for 20âmin at 4â°C to separate soluble and insoluble fractions, and fractions subsequently resuspended and diluted in lysis buffer. Sample fractions were mixed with SDS-loading buffer with 5âmM dithiothreitol, boiled and subject to immunoblot analysis.
For ex vivo IL-12p70 ELISA analysis, BALB/c mice bearing established hind-flank CT26 tumour were injected intratumourally with PBS, EcNcÎlon/ÎompT or EcNcÎlon/ÎompT/LLO+. Then 4â24âh after treatment, tumours were extracted and placed in ice-cold PBS containing HALT protease inhibitor cocktail without dimethylsulfoxide (DMSO) (ThermoFisher Scientific). Tumours were homogenized using a gentleMACS tissue dissociator (Miltenyi Biotec, C-tubes), and centrifuged at 3,000g for 10âmin at 4â°C. The supernatant was then collected and centrifuged at 10,000g for 20âmin at 4â°C to separate soluble and insoluble fractions. IL-12p70 in soluble sample fractions was analysed using the Mouse IL-12p70 Quantikine ELISA Kit (R&D systems) according to the manufacturerâs instructions.
Blood bactericidal assay
EcN wild-type or EcNcÎlon/ÎompT were cultured overnight in LB media without antibiotics. Cultures were centrifuged at 3,000g for 10âmin, resuspended in 1âml of ice-cold sterile PBS and normalized to OD600â=â1. Then 50âμl of OD600â=â1 microbe suspension was added to 1âml of single donor human whole blood (Innovative Research) in triplicate and incubated in a 37â°C stationary incubator. After 2âh of incubation, a sample was taken from each bloodâmicrobe mixture and serial dilution was prepared in PBS. Dilutions were plated on LB agar with erythromycin (25âμgâmlâ1). After incubation overnight at 37â°C, colonies were quantified by spot-forming assay and CFUâmlâ1 blood was calculated.
Biofilm assay
Biofilm formation assays were conducted as described previously73. Briefly, EcN wild-type, cryptic plasmid-cured (EcNc), Lon knockout (EcNcÎlon), OmpT knockout (EcNcÎompT) or double protease knockout (EcNcÎlon/ÎompT) were cultured for 48âh in LB media with 25âμgâmlâ1 erythromycin in borosilicate glass tubes in a 30â°C stationary incubator, with tube caps wrapped with parafilm to prevent evaporation. At 48âh, cultures were discarded and borosilicate tubes were washed three times with PBS. Tubes were inverted and allowed to dry for 6âh. Biofilms left on borosilicate tubes were stained with 0.1% (vol/vol) crystal violet for 15âmin. Crystal violet stain was discarded and tubes washed three times with PBS, then inverted and allowed to dry overnight. Crystal violet-stained biofilms were dissolved with 95% ethanol and transferred to 96-well plates for measurement of absorbance at 590ânm.
Phagocytosis assay
Bacterial phagocytosis assays were adapted from previous work74. Culture and isolation of murine BMDMs was performed as described previously75. Bulk femoral bone marrow cells from BALB/c or C57BL/6 mice were cultured on 15âcm non-treated cell culture Petri dishes in RPMI with 20% FBS, 25ângâmlâ1 M-CSF (R&D Systems) and 100âUâmlâ1 penicillinâstreptomycin. Media was replaced with fresh media after 4âdays of culture. After 7âdays of culture, plates were washed with PBS and adherent macrophages were dissociated using trypsin-EDTA. Macrophages were washed in PBS, resuspended at a density of 2âÃâ105 cells per ml in media and 1âml transferred to each well of 24-well plates. The 24-well plates were incubated overnight in a 37â°C incubator with humidified 5% CO2. EcN wild-type or EcNcÎlon/ÎompT, with or without a constitutive green-fluorescent protein (GFP)-expressing plasmid were cultured overnight in LB media with appropriate antibiotics. Bacterial cultures were centrifuged at 3,000g for 10âmin, washed three times with sterile PBS and resuspended at a density of 4âÃâ108 bacteria per ml in sterile PBS. Media from wells containing adherent macrophages was aspirated, wells washed three times with PBS and 1âml of RPMI with 5% mouse serum added to each well. Latrunculin A was added at a concentration of 1âμM to selected wells to inhibit phagocytosis. Next, 2âÃâ107 CFU of microbes were added to each well with each condition tested in triplicate. Microbial strains were incubated with BMDMs for 30âmin in a 37â°C incubator at 20ârpm. After 30âmin, media was aspirated and wells were washed six times with sterile ice-cold PBS. Adherent macrophages were dissociated using non-enzymatic cell dissociation buffer (Gibco), resuspended in fluorescence-activated cell sorting (FACS) buffer (PBS containing 2% FBS, 2âmM EDTA and 0.09% sodium azide) and analysed by flow cytometry.
In vitro BMDM activation
BMDMs were cultured as described above for phagocytosis assays. BMDMs were washed in PBS, resuspended at a density of 2âÃâ105âmlâ1 in media and 1âml transferred to each well of 24-well plates. The 24-well plates were incubated overnight in a 37â°C incubator with humidified 5% CO2. Wild-type EcN or EcNcÎlon/ÎompT with constitutive expression of OVA from a pUC origin plasmid were cultured overnight in LB media with appropriate antibiotics. Cultures were centrifuged at 3,000g for 10âmin, washed three times with PBS and resuspended at a density of 4âÃâ108 bacteria mlâ1 in sterile PBS. Media from wells containing macrophages was aspirated, wells were washed three times with PBS and 1âml of RPMI with 5% mouse serum was added to each well. Next, 1âÃâ107 live microbes were added to each well, with each condition replicated in triplicate. Live microbial strains were incubated with BMDMs for 6âh in a 37â°C incubator. After 6âh, media was aspirated and wells were washed six times with sterile ice-cold PBS. Adherent macrophages were dissociated using non-enzymatic cell dissociation buffer (Gibco), resuspended in FACS buffer and analysed by flow cytometry. DRAQ7 cell viability reagent was used to exclude dead cells (diluted 1:1,000 in FACS buffer). Extracellular antibodies for BMDM activation panel included CD80 (catalogue no. 16-10A1, Biolegend), MHC-II (catalogue no. M5/114.15.2, Biolegend), PD-L1 (catalogue no. 10F.9G2, Biolegend) and H2Kb-SIINFEKL (catalogue no. 25-D1.16, Biolegend), each used at 1:200 dilution.
In vitro BMDC stimulation
BMDC isolation and culture from mouse bone marrow was adapted from previous methods76. BMDCs from C57BL/6 mice were cultured on 15âcm non-treated cell culture Petri dishes in RPMI with 20% FBS, 20ângâmlâ1 GM-CSF (Biolegend) and 100âUâmlâ1 penicillinâstreptomycin. Every 1â2âdays for the first 4âdays, plates were gently washed and non-adherent granulocytes removed by aspirating 50% of the culture media with subsequent replacement of fresh media. On day 4, media was aspirated completely and replaced with fresh culture media with 20ângâmlâ1 GM-CSF. On day 6, BMDC plates were washed with PBS and loosely adherent and non-adherent cells collected. Cells were centrifuged at 300g for 5âmin, resuspended in fresh culture media and replated on 15âcm non-treated cell culture Petri dishes. On days 7â8, plates were washed with PBS and loosely adherent and non-adherent cells were collected. Cells were centrifuged at 300g for 5âmin, resuspended in fresh culture media at a density of 2.5âÃâ105 mlâ1 and 200âμl was transferred to 96-well plates and incubated overnight in a 37â°C incubator. The next day, media from wells containing BMDCs was aspirated, and 1âml of RPMI with 5% mouse serum was added to each well. BMDCs were pulsed with live bacteria at an multiplicity of infection of 10 for 2âh. Plates were centrifuged at 300g for 5âmin, media aspirated and replaced with fresh RPMI with 10% FBS, 10âμgâmlâ1 gentamicin and 100âUâmlâ1 penicillinâstreptomycin. Gentamicin concentration was increased to 40âμgâmlâ1 after 2â4âh. Plates were incubated for 5â48âh in a 37â°C incubator, at which time the supernatant was assessed for IL-12p70 using the Mouse IL-12p70 Quantikine ELISA Kit (R&D systems) according to the manufacturerâs instructions.
OT-I and OT-II T cell stimulation and proliferation
BMDCs were cultured as above, resuspended at a density of 2.5âÃâ105âmlâ1 and 5âÃâ104 BMDCs transferred to 96-well plates and incubated overnight in a 37â°C incubator. The next day, media from wells containing BMDCs was aspirated, and 1âml of RPMI with 5% mouse serum was added to each well. BMDCs were pulsed with 2âÃâ106 CFU of the respective bacterial strain for 2.5âh, plates were centrifuged at 300g for 5âmin, media aspirated and replaced with fresh RPMI with 10% FBS and 10âμgâmlâ1 gentamicin and 100âUâmlâ1 penicillinâstreptomycin. Gentamicin concentration was increased to 40âμgâmlâ1 after 2â4âh. Spleens from naive OT-I and OT-II mice were extracted, filtered through 100âµm cell strainers and washed in complete RPMI (RPMI-1640 supplemented with 10% (vol/vol) FBS, 1à GlutaMax, 1% (vol/vol) MEM non-essential amino acids solution (Gibco-11140050) and 100âUâmlâ1 penicillinâstreptomycin). OT-I and OT-II T cells were isolated from single-cell suspensions of spleens from the respective transgenic mouse using the EasySep Mouse T Cell Isolation Kit (StemCell Technologies) according to the manufacturerâs instructions. Purified OT-I and OT-II T cells were resuspended in T cell media (complete RPMI supplemented with 50âμM β-mercaptoethanol) at a density of 5âÃâ105âmlâ1 and 5âÃâ104 T cells incubated with 5âÃâ104 BMDCs pulsed with the respective microbial strains. For cytokine secretion assessment, T cells were incubated with BMDCs for 24âh, at which time supernatant was assessed for IFNγ and IL-2 using Mouse IFNgamma Quantikine ELISA Kit and Mouse IL-2 Quantikine ELISA Kit (R&D systems) according to the manufacturerâs instructions.
Carboxyfluorescein succinimidyl ester (CFSE) proliferation assays were conducted as previously described77. Here, 1âÃâ107 OT-I or OT-II T cells were resuspended in 1âml of room temperature PBS, and 1âμl of 5âmM CFSE (Biolegend) was added. T cells were incubated in CFSE solution for 5âmin at room temperature protected from light, after which time staining was quenched by adding ten times the staining volume of cell culture media. T cells were centrifuged at 300g for 5âmin, resuspended in T cell media at a density of 5âÃâ105âmlâ1 and incubated for an extra 10âmin at room temperature. Then 5âÃâ104âT cells were incubated with 5âÃâ104 BMDCs pulsed with the respective live microbial strains. At 48âh, 50% of the media from each well was gently aspirated so as to not disturb any cells, and replaced with fresh T cell media. At 72â96âh, OT-I and OT-II T cells were collected and analysed by flow cytometry. DRAQ7 cell viability reagent was used to exclude dead cells (diluted 1:1,000 in FACS buffer). Extracellular antibody staining for CFSE assays included antimouse CD3 (catalogue no. 17A2, Biolegend), used at 1:200 dilution.
Listeriolysin haemolytic activity assay
Sheep red blood cell (RBC) lysis by bacterial lysate was performed as described previously78. Briefly, bacteria were grown overnight in fresh LB containing appropriate antibiotics. Cultures were centrifuged at 3,000g for 10âmin, supernatants discarded and the cell pellet resuspended to OD600â=â8 in 0.1% (w/w) bovine serum albumin (BSA) in sterile PBS titrated to pH of 5.25 with 1âM HCl. Bacteria were sonicated for 2âmin. After sonication, the soluble fraction was isolated by centrifugation at 10,000g at 4â°C for 20âmin. Sheep RBCs were washed three times with PBS and resuspended at a final concentration of 6âÃâ108âmlâ1 in 0.1% (w/w) BSA in PBS titrated to pH of 5.25. Equal parts of bacterial lysate soluble fraction and sheep RBC suspension were mixed and incubated for 15âmin at 37â°C. After incubation RBC mixtures were centrifuged at 1,000g for 1âmin at 4â°C and supernatant absorbance at 541ânm was then measured to quantify RBC lysis.
Listeriolysin cytosolic access assay
BMDMs were cultured as described above for phagocytosis assays. BMDMs were washed in PBS, resuspended at a density of 2.5âÃâ105âmlâ1 in media, 100âml transferred to wells of an eight-well Lab-Tek Chamber Slide system (ThermoFisher) and incubated overnight in a 37â°C incubator. The next day, media from wells containing BMDMs was aspirated, and 1âml of complete RPMI without antibiotics was added to each well. BMDMs were then pulsed with 1.25 à 106 CFU of the respective bacterial strain for 60âmin. After the designated time media from each well was aspirated, wells were washed four times with PBS and media was replaced with fresh RPMI with 10% FBS and 40âμgâmlâ1 gentamicin and incubated in a stationary 37â°C incubator. After either 30 or 60âmin of more incubation, media was aspirated and wells washed four times with ice-cold PBS. Then 100âml of 100% methanol at â80â°C was then added to each well for fixation and allowed to incubate at room temperature for 10âmin. Methanol was then removed, 100âml of ice-cold PBS added to each well and slides incubated at 4â°C. Cells were permeabilized with 0.5% Triton X in PBS for 10âmin. Blocking solution in 10% heat-inactivated horse serum and 3% BSA was added to each well for 30âmin. After washing three times, primary antibodies in 1% heat-inactivated horse serum and 1% BSA were incubated overnight at 4â°C in a humidified chamber. The next day, slides were washed with PBS three times for 10âmin each and secondary antibodies were applied for 1âh at room temperature in the dark. DAPI (4â²6-diamidino-2-phenylindole) was applied as part of the secondary antibody cocktail for nuclear staining. Slides were washed in PBS three times before mounting coverslips with DAKO gel and stored at 4â°C until immunofluorescence analysis. Anti-ovalbumin (catalogue no. EPR27117-90, Abcam) and anti-CD11b (catalogue no. M1/70, Abcam) primary antibodies were used for staining, both at 1:200 dilution.
Animal experiments
All animal experiments were approved by the Institutional Animal Care and Use Committee (Columbia University, protocol AABQ5551). The 6â7-week-old female BALB/c, C57BL/6 and B6(Cg)-Tyrc-2J/J (Jackson Laboratories) mice were kept in accordance with all rules for animal research at Columbia University. Mice were housed in a facility with a 12âh lightâdark cycle, and provided unrestricted access to both food and water. The housing facility was maintained at 21â24â°C, and kept at 40â60% humidity. Sample size was determined on the basis of our previous studies and/or pilot experiments. For subcutaneous tumour models: 5âÃâ106 CT26 cells in 100âμl of sterile PBS were inoculated subcutaneously on the hind flank of BALB/c mice, or 5âÃâ105 B16F10 melanoma cells in 100âμl of sterile PBS subcutaneously on the hind flank (orthotopic) of C57BL/6 mice using a 26G needle on a 1âcc syringe. CT26 tumours were allowed to establish as indicated for each experiment, and mice were distributed between groups to equate the average starting tumour volume before treatment. B16F10 orthotopic tumours were allowed to establish for 9âdays, and initial average tumour volume equated between groups before treatment. Tumour dimensions were measured unblinded with a calliper every 1â3âdays for calculating tumour volumes using the equation (a2âÃâb)/2 (a is width, b is length, where width is the smaller dimension). Group tumour sizes were computed as meanâ±âs.e.m. Body weight was measured each time tumour measurements were taken. Animals were euthanized when any of the following criteria were met: tumour burden greater than 2âcm in the largest dimension for any subcutaneous tumour, greater than 20% body weight loss, as otherwise recommended by veterinary staff or when showing clinical signs of impaired health. To examine the requirement of individual T cell populations for the efficacy of the microbial neoantigen vaccines, mice were injected intraperitoneally with 200âμg (in 100âμl of InVivoPure pHâ6.5 buffer, BioXcell) of antimouse CD4 (clone GK1.5, BioXcell), 200âμg (in 100âμl of InVivoPure pHâ7.0 buffer, BioXcell) of antimouse CD8β (clone Ly-3.2, BioXcell) or 200âμg (in 100âμl of InVivoPure pHâ7.0 buffer, BioXcell) of IgG1 isotype control (clone HRPN, BioXcell) beginning 2âdays before the initiation of therapeutic treatment and every 2â3âdays thereafter until study endpoint.
In prophylactic vaccination studies, BALB/c mice received an intravenous injection of either EcNcÎlon/ÎompT/LLO+ OVA or EcNcÎlon/ÎompT/LLO+ nAg19 every 3â5âdays for a total of four injections. Four days after the final injection, 1âÃâ106 CT26 cells in 100âμl of sterile PBS were inoculated subcutaneously on the hind flank. In rechallenge studies, BALB/c mice that had cleared subcutaneous CT26 tumours on a single hind flank were engrafted with 1âÃâ106 CT26 cells on the opposite hind flank 100âdays after tumour clearance. Age-matched naive BALB/c mice were engrafted with 1âÃâ106 of the same CT26 cells on a single hind flank as controls.
For therapeutic studies in systemic metastases models, 5âÃâ105 CT26-Luc cells or 1.5âÃâ105 B16F10-Luc cells were injected in 100âμl of sterile PBS through the lateral tail vein with a 27G needle on 1âcc syringe. Metastases were allowed to establish for 4âdays in Balb/C mice before treatment for CT26-Luc, and for 2âdays in C57BL/6 albino mice (B6(Cg)-Tyrc-2J/J) for B16F10-Luc. Mice were randomly distributed between groups after metastases engraftment and before treatment. For in vivo luminescence tracking of metastases burden, mice were injected intraperitoneally with 125âμl of aqueous solution of d-Luciferin (50âmgâmlâ1) 6âmin before imaging, and placed under isoflurane anaesthesia for imaging using an in vivo imaging system (IVIS), with exposure time set to 6âmin. Total flux from the lungs (CT26-Luc) or body (B16F10-Luc) was used to quantify tumour burden. For evaluation of lung metastases burden at the timepoint of treatment initiation, mice were injected intraperitoneally with 250âμl of aqueous solution of d-Luciferin (50âmgâmlâ1) 6âmin before imaging and placed under isoflurane anaesthesia for imaging using an IVIS with exposure time set to 10âmin. After in vivo IVIS analysis, mice were then re-injected with 100âμl of aqueous solution of d-luciferin (50âmgâmlâ1) and lungs were extracted for ex vivo IVIS imaging with exposure time set to 2âmin.
No formal blinding was done for in vivo experiments. For all animal experiments, intratumoural treatments were injected directly into the tumour core with care to not allow leakage of any therapeutic solution. Intravenous treatments were injected through the lateral tail vein, with care not to allow leakage of any therapeutic solution.
SLP vaccination
The formulation and administration of SLP vaccines was adapted from previous studies15,79,80. Each dose contained either 20âμg of each 29-mer CT26 neoantigen peptide (19 neoantigens, 380âμg of total peptide per dose) and 50âμg of poly I:C in 200âμl of 10% DMSO/90% PBS (vol/vol) in Fig. 2e and Extended Data Fig. 5e,f, or 25âμg of each 29-mer CT26 neoantigen peptide (19 neoantigens, 475âμg total peptide per dose) and 100âμg of poly I:C in 200âμl of 10% DMSO /90% PBS (vol/vol) in Extended Data Fig. 5g. Therapeutic SLP vaccinations were administered subcutaneously to BALB/c mice with established hind-flank CT26 tumours on the contralateral hind flank using a 29G needle. SLP vaccine groups were treated on the same days as microbial therapeutic groups.
Ex vivo lung histology
Explanted lungs from BALB/C mice bearing CT26-Luc metastases or C57BL/6 albino mice (B6(Cg)-Tyrc-2J/J) bearing B16F10-Luc metastases were washed three times in PBS and placed in 10% formalin. After at least 24âh of fixation, lungs were transferred to 70% ethanol and subsequently embedded in paraffin. Then 50âμm consecutive sections were stained with haematoxylin and eosin. Lung sections were analysed for the presence of tumour foci.
Microbial administration for in vivo experiments
For therapeutic administration, bacterial strains were grown overnight in fresh LB media containing the appropriate antibiotics. Overnight cultures were centrifuged at 3,000g at 4â°C for 10âmin and washed three times with ice-cold sterile PBS. Microbes were delivered intratumourally at a concentration of 5âÃâ108âCFUâmlâ1 in sterile PBS, with 20âμl of injected using a 1âcc syringe with a 29G needle. For intravenous treatment, 100âμl of microbes were delivered at a concentration of 1âÃâ108âCFUâmlâ1 in sterile PBS, through the lateral tail vein using a 1âcc syringe with a 29G needle.
Biodistribution and in vivo bacterial dynamics
For biodistribution experiments, BALB/c mice bearing established hind-flank CT26 or lung metastatic CT26-Luc tumours were injected intravenously with 100âμl of 1âÃâ108âCFUâmlâ1 EcNcÎlon/ÎompT/LLO+. Then 96â120âh after a single i.v. injection for hind-flank tumours or at endpoint for lung metastases, tumours or tumour-bearing lungs and other organs were extracted from mice, weighed and homogenized using a gentleMACS tissue dissociator (Miltenyi Biotec, C-tubes). Homogenates were serially diluted in sterile PBS and plated on LB agar plates at 37â°C overnight. Colonies were quantified per organ and computed as CFU per gram of tissue (CFUâgâ1). For tracking bacterial colonization of subcutaneous tumours by microbial luminescence, tumour-bearing mice treated intratumourally or intravenously with wild-type EcN parental strain or genetic derivates were imaged using IVIS at various time points. For abscopal experiments, treated and untreated tumours were harvested 14âdays after a single intratumoural bacterial injection.
Ex vivo T cell killing assay
For B16F10-Luc specific killing, naive tumour-free C57BL/6 mice were injected intravenously every 4âdays with PBS, EcNcÎlon/ÎompT/LLO+ OVA or nAg42 for a total of four doses. Five days after the final dose spleens from treated mice were extracted, filtered through 100âµm cell strainers and washed in complete RPMI. T cells were isolated from single-cell suspensions of spleens from the respective mouse using the EasySep Mouse T Cell Isolation Kit (StemCell Technologies) according to the manufacturerâs instructions. Purified T cells were resuspended in T cell media for use in the specific lysis assay.
The luciferase-based killing assay was adapted from previous methods81. B16F10-Luc target cells were grown for 48âh in the presence of 100âUâmlâ1 murine IFNγ. Target cells were gathered and plated at 1âÃâ104 cells per well in a 96-well plate. After 12âh, T cells were added to each well to achieve designated effector-to-target ratios (10:1, 20:1 or 40:1). After 42âh of co-incubation, 50âUâmlâ1 IL-2 was added to all wells.
For CT26-Luc versus 4T1-Luc luciferase-based specific killing assay: BALB/c mice with established hind-flank CT26 tumours were treated intravenously with EcNcÎlon/ÎompT/LLO+ nAg19 on days 0 and 3. On day 8, tumours were extracted and mechanically homogenized, followed by digestion with collagenase A (1âmgâmlâ1, Roche) in isolation buffer (RPMI-1640 with 5% FBS, 1% l-glutamine, 1% penicillinâstreptomycin and 10âmM HEPES) with gentamicin (40âμgâmlâ1) for 1âh at 37â°C on a shaker platform at 150ârpm. Tumour homogenates were filtered through 100âµm cell strainers and washed in T cell media. Tumour-infiltrating T cells were isolated from single-cell suspensions of tumours from mice using the EasySep Mouse T Cell Isolation Kit (StemCell Technologies) according to the manufacturerâs instructions. Purified T cells were resuspended in T cell media.
CT26-Luc or 4T1-Luc target cells were grown for 12âh in the presence of 100âUâmlâ1 murine IFNγ. Target cells were gathered and plated at 1âÃâ104 cells per well in a 96-well plate. After 12âh, T cells were added to each well to achieve designated effector-to-target ratios (5:1 or 10:1), with 50âUâmlâ1 IL-2 added to all wells.
Luminescence from each well was quantified after addition of One-Glo Luciferase Assay System (Promega), as per the manufacturerâs instructions, after 24â96âh of coculture. Minimum lysis wells contained only the respective luciferase-expressing tumour target cells. In maximum lysis wells, 20âμl of media was replaced with 20âμl of 3% Triton X-100 60âmin before luminescence readout. Specific lysis (%) was calculated using the luminescence values of the respective conditions with the following formula: 100âââ(100âÃâ((sampleâââmaximum lysis)/(minimum lysisâââmaximum lysis))).
IFNγ ELISpot
BALB/c mice with established hind-flank CT26 tumours were treated intravenously with EcNcÎlon/ÎompT/LLO+ nAg19 on day 0 and 3. On day 8, tumours were extracted and mechanically homogenized, followed by digestion with collagenase A (1âmgâmlâ1, Roche) in isolation buffer (RPMI-1640 with 5% FBS, 1% l-glutamine, 1% penicillinâstreptomycin and 10âmM HEPES) with gentamicin (40âμgâmlâ1) for 1âh at 37â°C on a shaker platform at 150ârpm. Tumour homogenates were filtered through 100âµm cell strainers and washed in RPMI containing CTL-Wash Supplement (Immunospot) and 1% l-glutamine. Splenocytes from naive BALB/c mice were isolated in the same way, without digestion. T cells were isolated from single-cell suspensions of tumours from mice using the EasySep Mouse T Cell Isolation Kit (StemCell Technologies) according to the manufacturerâs instructions. Purified T cells were resuspended in CTL-Test Medium supplemented with 1% l-glutamine for use in the enzyme-linked immunosorbent spot (ELISpot) assay.
Mouse IFNγ Single-Color ELISpot plates and kits were purchased from Immunospot. ELISpot plates were prepared as per manufacturerâs instructions. Here, 5âÃâ105 naive splenocytes were plated with 2âÃâ104 TILs per well in 200âμl CTL-Test Medium supplemented with 1% l-glutamine and gentamicin (30âμgâmlâ1). Then 29-mer synthetic neoantigen or negative control (OVA) peptides were added to each well at a final concentration of 5âμgâmlâ1. Cells were stimulated overnight in a stationary 37â°C incubator with atmosphere of humidified 5% CO2. After incubation, plates were developed as per the manufacturerâs protocol and spots quantified using a CTL Immunospot S6 Universal machine and CTL ImmunoSpot software v.7.0.24.0.
Flow cytometry immunophenotyping
For CT26 flow-cytometric immunophenotyping, BALB/c mice with hind-flank CT26 tumours received intravenous treatment with the indicated microbial therapeutic or PBS on day 0. Two or 8âdays after treatment, TDLNs and/or tumours were extracted. For B16F10 flow-cytometric immunophenotyping, C57BL/6 mice with hind-flank, orthotopic B16F10 tumours received intravenous treatment with the indicated microbial therapeutic or PBS on day 0 and 3. Eight days after treatment, tumours were extracted. Lymphoid and myeloid immune subsets were isolated from tumour tissue by mechanical homogenization of tumour or TDLN tissue, followed by digestion with collagenase A (1âmgâmlâ1, Roche) and DNase I (0.5âµgâmlâ1, Roche) in isolation buffer (RPMI-1640 with 5% FBS, 1% l-glutamine, 1% penicillinâstreptomycin and 10âmM HEPES) for 1âh at 37â°C for tumours or 30âmin at 37â°C for TDLNs, on a shaker platform at 150ârpm. For ex vivo lymphocyte stimulation with PMA and ionomycin, TDLNs were not digested beforehand. Tumour and TDLN homogenates were filtered through 100âµm cell strainers and washed in isolation buffer. To measure overall cytokine production by T cells, cells were stimulated for 3âh with PMA (50ângâmlâ1, Sigma-Aldrich) and ionomycin (1ânM, Calbiochem) in the presence of brefeldin A (1âμgâmlâ1). To measure neoantigen-specific cytokine production by T cells, cells were stimulated for 5âh with pools of peptides (2âμgâmlâ1) representing the neoantigens encoded in therapeutic strains in the presence of brefeldin A (1âμgâmlâ1). Cells were stained in FACS buffer. Ghost Dye cell viability reagent was used to exclude dead cells (diluted 1:1,000 in PBS). Extracellular antibodies for lymphoid immunophenotyping included: CD4 (RM4-5, Biolegend), NKp46 (29A1.4, BD Biosciences), NK1.1 (PK136, Biolegend), CD45 (30-F11, BD Biosciences), B220 (RA3-6B2, BD Biosciences), CD19 (6D5, Biolegend), CD8a (53-6.7, Biolegend), TIM-1 (RMT1-4, BD Biosciences) and CD69 (H1.2F3, BD Biosciences). After extracellular staining, cells were washed with FACS buffer, and fixed using the FOXP3/transcription factor staining buffer set (Tonbo), as per the manufacturerâs instructions. Intracellular antibodies for lymphoid immunophenotyping included: Foxp3 (FJK-16s, Thermo), CD3ε (145-2C11, Biolegend), TCRβ (H57-507, BD Biosciences), Ki-67 (SolA15, Thermo), Granzyme-B (QA16A02, Biolegend), TNF (MP6-XT22, Biolegend) and IFNγ (XMG1.2, Biolegend). For myeloid immunophenotyping, extracellular antibodies included: Ly6C (HK1.4, Biolegend), I-A/I-E (M5/114.15.2, BD Biosciences), XCR1 (ZET, Biolegend), CD11b (M1/70, Biolegend), CD103 (2E7, Biolegend), CD45 (30-F11, BD Biosciences), F4/80 (BM8, Biolegend), CD11c (HL3, BD Biosciences), CD172a/SIRPα (P84, Biolegend), Ly6G (1A8, Biolegend and BD Biosciences), PD-L1 (10âF.9G2, Biolegend), CD301b (URA-1, Biolegend), CD3 (145-2C11, Biolegend), CD19 (1D3, Biolegend), NK1.1 (PK136, Biolegend), NKp46 (29A1.4, Biolegend) CD64 (X54-5/7.1, Biolegend), CD80 (16-10A1, Biolegend) and CD86 (GL-1, BD Biosciences). All antibodies for flow cytometry were used at a 1:200 dilution. After staining, cells were washed and resuspended with FACS buffer for flow cytometry analysis using a BD LSRFortessa or Cytek Aurora cell analyser. FACS Diva or SpectroFlo software was used for data acquisition. Collected flow cytometry data were analysed using FlowJo.
Synthetic peptides
Synthetic peptides representing neoantigens for lymphocyte restimulation assays and vaccination were synthesized by and purchased from Peptide v.2.0. All peptides were above or equal to 95% purity.
Statistics and reproducibility
Statistical analyses and P value calculations were performed using GraphPad Prism v.9 and v.10. For each experiment, the particular statistical analysis is detailed in the respective figure legend. A two-tailed unpaired Studentâs t-test, one-way analysis of variance (ANOVA) or two-way ANOVA with appropriate post hoc test was used for data that were roughly normally distributed. For analysis of KaplanâMeier survival experiments, the log-rank (MantelâCox) test was used. All analyses were two-tailed. For all statistical analyses, NS denotes not significant, which is Pâ>â0.05.
For Fig. 4a and Extended Data Fig. 7b, immunoblot data are representative of four independent experiments. In Extended Data Figs. 1b and 2h, immunoblot data are representative of three independent experiments. Immunofluorescence data in Extended Data Fig. 2i are representative of three independent experiments. Histology data in Extended Data Figs. 5h and 8f are representative of three independent experiments. All other results in the paper were replicated at least two to three times in independent experiments.
Biological materials availability
Reasonable requests for biological materials used in this study will be promptly reviewed by Columbia Technology Ventures to verify whether the request is subject to any intellectual property or confidentiality obligations. Any materials that can be shared will be released through a material transfer agreement.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.