Patients and treatment interventions
The patients included in this analysis were identified through a retrospective review of all patients with recurrent, platinum-resistant or refractory OCCC (including clear cell carcinomas arising from peritoneal and fallopian tube origins), defined as progression within 180 days of a previous line of platinum-containing therapy, treated with ICB (MD Anderson Cancer Center Institutional Review Board (IRB) 2020-1098). The patients included in this analysis were treated on two different clinical trial protocols, both IRB approved by MD Anderson, in which combination CTLA-4 and PD-1/L1-targeting ICB was administered (Clinicaltrials.gov: NCT03026062 (n = 33) and NCT01928394 (n = 1)). Informed consent was obtained from all of the patients enrolled in the two trials before starting treatment. NCT03026062 was an open-label, adaptively randomized phase II trial of tremelimumab and durvalumab administered in combination or sequentially. Sequential therapy included tremelimumab 3 mg per kg every 4 weeks for up to 4 doses, followed by durvalumab 1.5 g every 4 weeks for up to 9 doses after progression. Combination therapy consisted of tremelimumab 1 mg per kg plus durvalumab 1.5 g every 4 weeks for up to 4 doses followed by durvalumab monotherapy (1.5 g for up to 9 doses or until progression). The outcomes for the non-OCCC histology cohorts were reported previously48. Notably, enrolment on this trial was amended, initially limiting it to patients with OCCC, and later further restricting participation to those with OCCC with inactivating PPP2R1A or activating AKT alterations. An additional patient included in this study received treatment on NCT01928394, an open-label phase II trial in which the patients were stratified by platinum-free interval and then randomized to different dosing schedules of nivolumab and ipilimumab. The participant included in this study received ipilimumab 1 mg per kg every 6 weeks plus nivolumab 3 mg per kg every 2 weeks until progression.
Clinicopathological data collection and analysis
Demographic and clinicopathological information, including tumour somatic mutation testing and mismatch repair/microsatellite stability status, was collected from the medical record. Tumour mutation testing using next-generation sequencing was obtained as part of the patients’ standard care either at MD Anderson Cancer Center or through a commercial lab (typically Caris or FoundationOne). Clinical DNA mutation testing was not available for two participants; in these cases, T200 next-generation sequencing was performed on their pretreatment biopsies as previously described49.
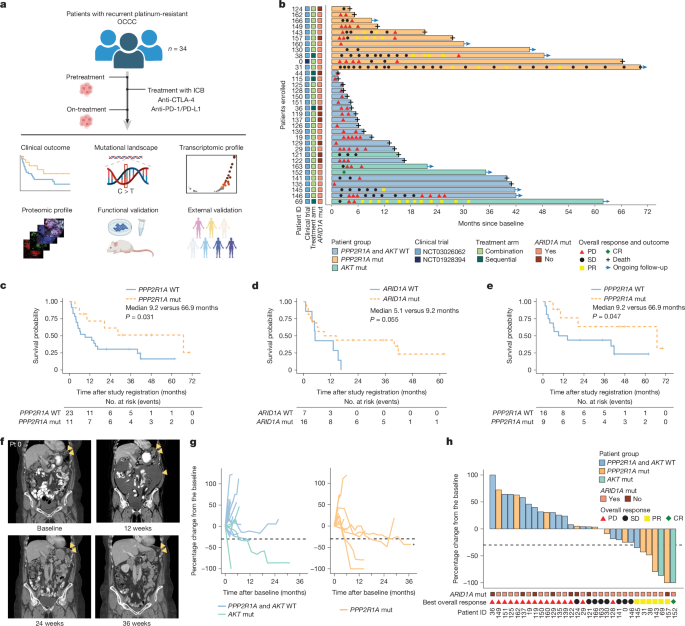
RECIST v.1.1, modified for immunotherapy50, was used according to the respective clinical trial protocols to assess the response. Independent radiology assessment was performed by experienced radiologists at The University of Texas MD Anderson Cancer Center, and progressive disease was determined on the basis of both the target lesion size as well as non-target lesion equivocal progression. The patients in the study were allowed to continue treatment beyond progression based on risk–benefit considerations. A swimmer plot was used to describe patient overall responses and outcomes. Spider plots and waterfall plots were used to describe target lesion responses, as revealed by the sum of tumour diameters, during the on-trial period. OS was calculated from the time of study registration to the earliest date of death or data cut-off (29 January 2024). PFS was calculated from the time of study registration to the date of disease progression or death or the last time of clinical evaluation. For patients 44, 115 and 126, owing to rapid disease progression leading to the patients’ death, no tumour measurements were performed based on CT scan, and PFS was determined based on clinical criteria. OS and PFS were estimated using the Kaplan–Meier method and compared by variables of interest using one-sided log-rank rest. Within the cohort of patients with somatic PPP2R1A mutations, the patients were further dichotomized into long-term versus short-term survivors on the basis of the median OS for further translational analyses. Clinical data analysis was performed in R statistical environment (v.4.3.1; https://www.r-project.org/).
Clinical sample collection
Pretreatment image-guided core biopsies, stored as formalin-fixed paraffin-embedded (FFPE) samples, were collected before the first dose of the study drug. On-treatment biopsies, also stored as FFPE samples, were performed when there was a lesion amenable to biopsy as assessed by the cycle 2 imaging study by an MD Anderson Cancer Center radiologist. When possible, on-treatment biopsies were obtained from the same area of tumour as the pretreatment biopsies. Samples were collected, labelled, processed and stored by the MD Anderson Cancer Center Gynecologic Oncology Tumour Bank for translational studies. Sample identification and associated patient-level data were entered into a secure database.
mRNA library preparation and sequencing
Stranded mRNA libraries were prepared using the KAPA Stranded mRNA-seq Kit (Roche). In brief, poly(A) RNA was captured from 10–250 ng of total RNA using magnetic oligo-dT beads. After bead elution and clean-up, the resultant poly(A) RNA was fragmented using heat and magnesium. First-strand synthesis was performed using random priming followed by second-strand synthesis with the incorporation of dUTP into the second strand. The ends of the resulting double-stranded cDNA fragments were repaired, 5′-phosphorylated, 3′-A-tailed and Illumina specific indexed adapters were ligated. The products were purified and enriched for a full-length library with 9–16 cycles of PCR. The strand labelled with dUTP is not amplified, resulting in a strand-specific library. The libraries were quantified using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific) and assessed for size distribution using the 4200 Agilent TapeStation system (Agilent Technologies) according to the manufacturer’s instructions. Equimolar quantities of the indexed libraries were then multiplexed 28 libraries per pool. The library pool was quantified by quantitative PCR, then sequenced on one lane of the NovaSeq 6000 (Illumina) S4 flow cell using the 100-nucleotide paired-end run format.
RNA-seq data processing and analysis
RNA-seq FASTQ files were processed through FastQC (v.0.11.5), a quality-control tool to evaluate the quality of sequencing reads at both the base and read levels, and RNA-SeQC (v.1.1.8) to generate a series of RNA-seq-related quality-control metrics51. All of the samples passed the quality checks for this study. STAR 2-pass alignment (v.2.7.0 f)52 was performed with the default parameters to generate RNA-seq BAM files. HTSeq-count (v.0.11.0)53 was applied to aligned RNA-seq BAM files to count for each gene how many aligned reads overlap with its exons. In the initial assessment of transcriptomic-level similarity and distinctions of all of the samples, two samples shown as significant outliers were removed from further analysis. Differential gene expression analyses were then performed using the R package DESeq2 (v.1.42.1)54, with raw count data as input. A ranked list of coding genes was generated based on the Wald statistic in DESeq2, and subsequently processed by GSEA55 using the R package clusterProfiler (v.4.6.2)56 against the Hallmark gene sets from Molecular Signature Database (MSigDB)57 to identify significantly enriched pathways. The false-discovery-rate-corrected q values were used for multiple-hypothesis-testing adjustment58. The HTSeq raw count data were then normalized to transcripts per kilobase million (TPM), and the TPM expression values were used as an input to the software CIBERSORTx59 (https://cibersortx.stanford.edu/) for immune cell deconvolution in each sample, using the default signature matrix LM22. Next, intergroup comparisons of the abundance of certain immune cell populations were performed using Wilcoxon rank-sum and signed-rank tests. The results from above analyses were visualized using the R package ggplot2 (v.3.4.2). All of the downstream analyses were performed in the R statistical environment (v.4.3.1; https://www.r-project.org/).
TCR and BCR repertoire analysis
The T cell and B cell receptor repertoires were reconstructed using TRUST4 (v.1.0.5.1)60 on the aligned RNA-seq BAM files. Repertoire richness was calculated as the total number of unique receptor sequences with >1 mapped reads, to account for singletons. Downstream data analysis was performed with the R package immunarch (v.1.0.0; https://immunarch.com/). The TCR and BCR richness was compared between PPP2R1A-mutant and wild-type samples. Only samples with detectable TCR/BCR were used for comparisons. Wilcoxon signed-rank and rank-sum tests were performed for comparing paired samples and the entire cohort, respectively.
Multiplexed protein imaging and data analysis
A multiplexed barcoding image analysis platform was performed using methods and reagents similar to those previously described61. Using the PhenoCycler Fusion platform (Akoya Biosciences) staining kit (Akoya, 7000008), the antibody cocktail was prepared by combining staining buffer, N blocker, G blocker, J blocker, S blocker and all barcoded antibodies, including pan-cytokeratin, CD45, CD3e, CD8, CD4, CD45RO, granzyme B, CD56, CD20, CD19, CD21, FOXP3, PD-1, PD-L1, CTLA-4, OX40, CD73, CD39, CD68, ARG1, CD206, CD31, HLA-DR, Ki-67, CD33, MHC-I and 4′,6-diamidino-2-phenylindole (DAPI) (detailed information of the antibody panel is provided in Supplementary Table 5). The slides were placed in a plastic humidity chamber, and 190 µl of antibody cocktail, including all of the markers above, was dropped onto the sample and incubated at 4 °C overnight. Next, a 96-well reporter plate (Akoya, 7000006) was prepared containing barcoded fluorophores according to custom-designed multiplex barcoding experiments. Each well was filled with 245 µl of a solution containing the reporter and specific barcode fluorophores for that cycle and placed into the PhenoCycler Fusion instrument (Akoya Biosciences). Each cycle will have 2 or 3 antibodies plus their specific barcode fluorophores and DAPI. At the end of each cycle, each image from the tissue sample was captured at a high magnification of ×20 using the PhenoCycler Fusion fluorescence microscope (Akoya Biosciences). Then, the signal was erased to start a new cycle until the desired antibody image panel was complete. Experienced pathologist analysed each tissue sample using image analysis software (QuPath v.0.4.4)62. For image analysis, the whole section of the tissue was divided into a tumour compartment, characterized by the nets of tumour cells, and the stroma compartment, the tissue between the tumour nets; and then the individual cell boundaries were determined using the pretrained StarDist algorithm within QuPath63. Marker co-expression was used to identify specific cell phenotypes in the tumour and stroma compartments. The data were preprocessed to be consolidated using the R package phenoptr (v.0.2.2, Akoya Biosciences). The densities of cell phenotypes (marker co-expression) were expressed as the number of cells per mm2. The cell densities were then compared between PPP2R1A-mutant and wild-type PPP2R1A samples using two-sided Wilcoxon rank-sum tests. Multicellular neighbourhood analysis was performed, with each multicellular neighbourhood defined as a collection of cells of which the centroids were within 80 μm of the centre cell. The majority of multicellular neighbourhoods contained 50–200 cells (mean, 116 cells; median, 99 cells), which aligns with recent studies22,23,24. Each cell in the images was taken as the centre iteratively to obtain a neighbourhood composition vector. In such a way, a neighbourhood composition matrix was constructed for the whole dataset. The neighbourhood compositions of certain cell types were then compared between different timepoints or sample groups using two-sided Wilcoxon rank-sum tests.
Establishment of genetically modified cell lines
To generate PPP2R1A knockdown cell lines, SKOV3 cells were transduced with retrovirally expressed shRNAs. Fully synthesized dsDNA fragments (Twist Bioscience) encoding gene-specific shRNAs were inserted into the pSuper.retro.puro (VEC-PRT-0002, OligoEngine) according to the manufacturer’s protocol. To generate retroviral supernatants, HEK293T cells were seeded 16 h before transfection and transfected with the retroviral vectors encoding different shRNAs (Supplementary Table 6), along with lentiviral packaging plasmids, pCMV-VSV-G and pBS-CMV-GagPol (8454 and 35614, Addgene) by the jetPRIME transfection reagent (101000046, VWR) according to the manufacturer’s protocol. Viral supernatants were collected at 48 h after transfection and filtered by 0.45 μm PVDF Syringe Filter Unit (SLHV033NK, Millipore-Sigma) to remove cell debris. Designated titres of retrovirus were used to infect cells in the presence of 8 μg ml−1 hexadimethrine bromide (107689, Sigma-Aldrich). A series of genetic modified cell lines was generated by sorting GFP+ cells 72 h after retrovirus transduction. Cells transduced with shRNA with the non-target control were used as negative-control cells. Subsequent immunoblot assays were performed to assess the knockdown efficacy.
To ectopically express wild-type PPP2R1A and the P179R mutant in SKOV3 cells, dsDNA fragments encoding human open reading frames (ORFs) of wild-type PPP2R1A (NM_014225.6) and the P179R mutant were inserted into the lentiviral vector, pLVX-IRES-ZsGreen1 (PT4064-5, Takara Bio). To generate lentiviral supernatants, HEK293T cells were seeded 16 h before transfection and transfected with the lentiviral vectors encoding wild-type PPP2R1A and P179R mutant, along with the lentiviral packaging plasmids pCMV-VSV-G and psPAX2 (8454 and 12260, Addgene), using the jetPRIME transfection reagent according to the manufacturer’s protocol. The viral supernatants were collected 48 h after transfection and filtered as previously described. Designated titres of lentivirus were used to infect cells in the presence of 8 μg ml−1 hexadimethrine bromide (Sigma-Aldrich). Stable cell lines were generated by sorting GFP+ cells 72 h after lentivirus transduction. Cells transduced with empty vector pLVX-IRES-ZsGreen1 were generated and used as control cells.
CRISPR–Cas9-mediated PPP2R1A gene editing
To perform adenine base editing (ABE) targeting the c.547C>T mutation of the PPP2R1A gene in HEC50B cells, the primer for the single guide RNA (sgRNA) was designed as TGCGGCCCGCCGCACCATGG and was synthesized by IDT. The sgRNA was cloned into pLentiCRISPR V2 (Addgene, 83480). ABE8e (Addgene, 138489) was used for ABE64. To remove Cas9 from the vector, the pLentiCRISPR V2 vector was digested with XbaI and BamHI. A PCR fragment for ABE8e containing an overlapping cloning site with the digested vector was inserted into the vector using the ClonExpress Ultra One Step Cloning kit (Cellagen C115-01). The following oligonucleotides were used for PCR: forward, 5′-GAACACAGGACCGGTTCTAGAGCCACCATGAAACGGACAG-3′; reverse, 5′-AAGTTTGTTGCGCCGGATCCGACTTTCCTCTTCTTCTTGGGCTCGA-3′. The resulting plasmid was used for lentivirus packaging and infection. Single clones were then selected and validated for editing efficiency by Sanger sequencing. To introduce R183Q (c.548G>A mutation) targeting the PPP2R1A gene in OVCAR429 cells, cytosine base editing (CBE) was used. The primer for the sgRNA was designed as CTGCGGCCCGCCGCACCATG and was synthesized by IDT. The sgRNA was cloned into lentiGuide-puro (Addgene, 52963). For CBE, hyBE4max (Addgene, 157942)65 was used and cloned into pLentiCas9-blast (Addgene, 52962) using the same cloning protocol mentioned above. The resulting plasmids were used for lentivirus packaging and infection. Single clones were then selected and validated for editing efficiency by Sanger sequencing. Notably, the CBE induced two mutations, c.548G>A and c.549G>A, ultimately leading to the amino acid change from Arg (CGG) to Gln (CAA).
Furthermore, we also generated HEC50B and OVCAR429 overexpressing hCD19 for downstream cytotoxicity assays using a similar approach to that described previously66. In brief, the hCD19 ORF was obtained from the Wistar Institute Molecular Screening and Protein Expression Facility, and then PCR-amplified and cloned into pLVX-M-puro vector (Addgene, 125839). HEK293T cells were transfected by Lipofectamine 2000 for lentivirus packaging. Lentivirus was collected and filtered with 0.45 mm filter 48 h after transfection. Cells infected with lentivirus were selected in 1 μg ml−1 puromycin or 1 μg ml−1 blasticidin 48 h after infection. The cells were then analysed using flow cytometry. Cells were washed with PBS containing 0.5% (w/v) BSA (PBS/BSA). APC anti-human CD19 antibody (BioLegend, 302212) was used at a 1:20 dilution. After staining, cells were then washed twice with cold PBS/BSA buffer, resuspended in this buffer and analysed using flow cytometry.
To generate R183Q (c.548G>A mutation) targeting the Ppp2r1a gene in Arid1a−/−Pik3caH1047R mouse ovarian clear cell cancer cells, CBE was performed using the sgRNA 5′-CAGCGGCCCGCCGCACCATG-3′ synthesized by IDT. In brief, the sgRNA was cloned into sgRNA-expressing vector containing a mCherry marker (Addgene, 210212). hyBE4max-eGFP (Addgene, 157942) and sgRNA expressing plasmid (10 μg) were co-transfected into 6 cm dish using polyethylenimine (MedChemExpress, HY-K2014). After 72 h, transfected cells were enriched by fluorescence-activated cell sorting through the selection of positive eGFP and mCherry expression. Single clones were then selected and validated for editing efficiency by Sanger sequencing.
Immunoblot
Cells were lysed with RIPA buffer (Thermo Fisher Scientific, 89901) with protease/phosphatase inhibitor (Thermo Fisher Scientific, A32959) on ice for 30 min. Proteins were denatured using SDS loading buffer (Bio-Rad, 1610747) at 95 °C for 5 min. Proteins were then separated by SDS–PAGE gel and transferred to a nitrocellulose membrane (Bio-Rad, 1620115). The membranes were blocked with 5% non-fat milk and incubated with primary antibodies at 4 °C overnight. The following antibodies were used: rabbit anti-AKT1 (Cell Signaling, 4691, 1:1,000), rabbit anti-phospho-Thr308-AKT1 (Cell Signaling, 4056, 1:1,000), rabbit anti-GAPDH (Cell Signaling, 5174, 1:3,000), rabbit anti-PPP2R1A (Cell Signaling, 2041, 1:1,000) and mouse anti-β-actin (Cell Signaling, 3700, 1:3,000). Objective signals were amplified with HRP-conjugated secondary antibodies (Cell Signaling, 77074) and detected by chemiluminescent substrate (Bio-Rad, 34094). The intensity of protein bands was detected using the ChemiDoc Imaging System (Bio-Rad) and quantified using the Image Lab software (v.5.2.1 build 11).
Human CAR-T preparation
Human T cells were activated by priming human CD3+ T cells isolated from PBMCs of the healthy donor with human T-Activator CD3/CD28 Dynabeads (11161D, Gibco) and 200 U ml−1 IL-2 (rhIL-2; Prometheus Laboratories, NDC Code 65483-116-07) for 24 h. During the activation, T cells were transduced with human B7H3-(SS1)-hBBZ-CAR encoding g-retrovirus to make T cells targeting human B7H3 in the presence of 10 μg ml−1 protamine sulfate (P3369, Millipore-Sigma) by centrifugation for 2 h at 1,800 rpm at room temperature. Transduction rates of CAR vectors were >85%.
The construction of hCD19 CAR-T was performed similarly to as described previously66. In brief, the FMC63 mouse-derived anti-human CD19 single chain variable region67 was synthesized by Genscript and cloned into the pTRPE lentiviral expression plasmid backbone68. An EF1a promoter drives the expression of a GFP reporter upstream of a T2A cleavage site, followed by the chimeric antigen receptor consisting of the FMC63 mouse anti-human single chain variable region followed by the CD8a hinge, CD28 transmembrane domain, the CD28 intracellular costimulatory domain and the CD3z chain.
Cytotoxicity assays
SKOV3 cells were prestained with CFSE (565082, BD Bioscience) according to the manual and seeded into the 96-well plate with round-bottom wells before the LB-100 (S7537, Selleck Chemicals) treatment. Tumour cells were mixed with paired B7H3 CAR-T cells at an effector:target ratio of 1:1 at 37 °C for 3 h. The percentage of activated caspase-3+ cells in tumour cells, stained with active caspase-3 antibody (C92-605, 560626, BD Bioscience), was assessed using LSRFortessa X-20 Cell Analyzer (BD Biosciences) and used to determine the tumour apoptosis rate induced by tumour-reactive T cells.
A set of genetically modified SKOV3 cell lines was plated into six-well plates (2 × 105 cells per well in completed medium). The next day, the 6-day cultured B7H3 CAR-T cells were co-cultured with SKOV3-derived cells at an effector:target ratio of 1:1 overnight. Next, dead cells and T cells were gently washed twice with prewarmed PBS. Adherent cells were collected using 0.25% trypsin-EDTA and stained with Trypan Blue for manual cell counting. The killing efficiency was calculated as (1 − number of live cells in T cell treated group/number of live cells in control group) × 100%.
For the cytotoxicity assay of the HEC50B and OVCAR429 cell lines, 10,000 target cells were seeded into 24-well tissue culture plates. hCD19 CAR-T cells were co-cultured with target cells at each effector:target ratio in RPMI 1640 fully supplemented in the absence of cytokines. For treatment, the cell medium was changed every 2 days with appropriate CAR-T cells for a total of 4 days. Next, colonies were stained with 0.05% crystal violet and the integrated density was measured using NIH ImageJ (v.1.54g).
Humanized PDXs of endometrial cancer
The generation of humanized-bone marrow, liver and thymus (Hu-BLT) mice69,70 was performed at The Wistar institute in accordance with Institutional Animal Care and Use Committee (IACUC)-approved protocols (201360 and 201533). In brief, 6-to-8-week-old female NSG mice (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ, Jackson Laboratory) were housed and maintained in individual microisolator cages in a rack system capable of managing air exchange with filters. The room temperature, humidity and pressure were controlled. Mice were maintained at 22–23 °C with 40–60% humidity and under a 12 h–12 h light–dark cycle. All of the mice were pretreated by busulfan at 30 mg per kg and were then implanted with human fetal thymic tissue fragments and fetal liver tissue fragments under the mouse renal capsule. After surgery, the mice were injected through the tail vein with CD34+ haematopoietic stem cells isolated from human fetal liver tissues. Human fetal liver and thymus tissues were procured from Advanced Bioscience Resources with The Wistar Institute IRB 21809310.
PDXs were generated using human endometrial tumour samples obtained from Christiana Care under IRB CCC-41045. HLA-A genotyping was performed with fetal liver and PDX tumour. In brief, genomic DNA of human tumour and fetal tissues was extracted using the Monarch Genomic DNA Purification Kit (NEB, T3010S). The HLA-A locus sequence was determined using the Olerup SSP HLA-A genotype kit (CareDx, 101.401-48). The third passage of the collected PDX tissue was transplanted subcutaneously into the Hu-BLT mice under aseptic conditions. Tumour samples were prepared by cutting endometrial PDX tumour chunks into small pieces of around 1 × 2 × 2 mm. Then, 2 weeks after PDX implantation, the mice were randomly assigned into two treatment groups: anti-PD-L1 antibody (Selleck, A2013) and human IgG1 isotype control (Syd Lab, PA007125) (4 mg per kg body weight in 100 μl PBS intraperitoneally, twice a week). After 3 weeks of treatment, the mice were euthanized 1 week later, and the tumours were surgically dissected. The tumour burden was assessed based on tumour weight.
Syngeneic models of OCCC
The study protocol was approved by The University of Texas MD Anderson Cancer Center (protocol number, 00002399-RN00). For syngeneic models, 2 × 106 mouse OCCC cells were subcutaneously injected into 6-to-8-week-old female immunocompetent C57BL/6 mice (027, 2159769, Charles River Laboratories). Mice were maintained under the same housing conditions as the PDX models, as described above. Then, 9 days after injection, the mice were randomly assigned into two treatment groups: anti-mouse PD-L1 antibody (BioXcell, BE0101) or an IgG2b isotype control (BioXCell, BE0090) (2.5 mg per kg body weight in 100 μl PBS intraperitoneally, once a week). The body weight and tumour size were measured twice a week. After 2 weeks of treatment, the mice were euthanized and the tumours were surgically dissected. The tumour burden was assessed based on tumour weight.
IACUC guidelines of The Wistar Institute and The University of Texas MD Anderson Cancer Center were followed in determining the time for ending survival, such as when the tumour burden exceeded 10% of the body weight or the tumour size exceeded 2 cm in any direction. The limits were not exceeded in both the PDXs and the syngeneic mouse models.
Uterine cancer validation cohort
Pharmacy records were queried in November 2023 to identify patients with uterine or endometrial cancer who received len–pem combination therapy over the past four years (from September 2019 to November 2023). Demographic and clinicopathological data, including cancer histology, mismatch repair/microsatellite stability status and tumour somatic mutation testing, were abstracted from the medical record under MD Anderson IRB approved protocol 2020-1098. The following histologies were included: serous, clear cell, carcinosarcoma, mesonephric-like adenocarcinoma and mixed high-grade carcinoma, given the relatively higher prevalence of PPP2R1A mutations in these high-risk histologies. The patients were also stratified by TP53 status, given that most OCCCs are wild-type TP53, while most high-grade endometrial carcinomas are TP53-mutated. Patients who never actually received len–pem, those with MSI-H/dMMR tumours and those who had no available imaging after starting treatment were excluded. PFS was calculated from the treatment start date to date of radiographic progression or last imaging study (if no progression). Patients who stopped treatment owing to toxicity were censored at the time of initiation of a new line of therapy. OS was estimated using the Kaplan–Meier method and compared by PPP2R1A mutation status using two-sided log-rank tests.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.