Phylogenetic analysis and multiple sequence alignments
A list of the sequences of CREs used for phylogenetic analysis is provided in Supplementary Table 3. Phylogenetic trees were generated using Geneious Tree Builder (Geneious software v.11.1.5): global alignment with free end gaps; Blosum62 as cost matrix; Jukes–Cantor distance model; method neighbor-joining; gap open penalty 10; and gap extension penalty 0.2. For multiple-sequence alignments (MSAs), Geneious Alignment with the same parameter, but Gap open penalty 20 and 5 refinement iterations was used. Pairwise identity within and between Metazoan/Apicomplexan proteins was calculated from MSAs and kernel smoothing was applied.
Nucleosome reconstitution
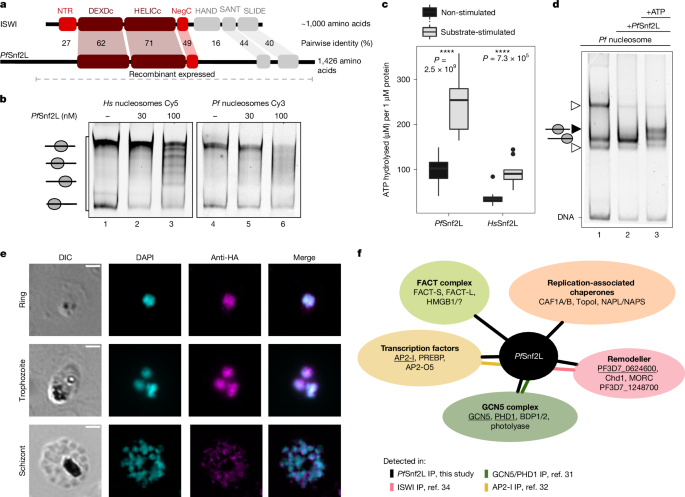
DNA templates for nucleosome assembly were synthesized by PCR, using (Cy3-/Cy5-labelled) oligonucleotides binding to the flanking regions of the 147 bp NPS, derived from the nucleosome assembly 601 sequence, creating various linker lengths (Extended Data Fig. 2b): 0-NPS-0, 6-NPS-47, 77-NPS-7747. Analogously, plasmodial DNA templates were amplified from genomic DNA. Recombinant histones—canonical human and P. falciparum and variant P. falciparum (H2A.Z/B.Z containing)—were expressed, purified and refolded as previously described8. Bos taurus and Gallus gallus histone octamers were purified from calf thymus or chicken blood, respectively, as described previously48,49 (Extended Data Fig. 2c). Nucleosome assembly was performed using the salt dialysis method. Histone octamers and DNA were mixed in 40–200 µl high-salt buffer (10 mM Tris pH 7.6, 2 M NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, 0.05% Igepal CA-630) supplemented with 200 ng µl−1 BSA in small dialysis chambers. High salt concentrations were reduced to 200 mM NaCl overnight at 4 °C. Reconstituted nucleosomes were analysed by gel electrophoresis on 6% polyacrylamide gels in 0.4× TBE buffer and visualized by fluorescence scanning (Typhoon, FLA-9500) or ethidium bromide staining (Extended Data Fig. 2d,g–l). Raw scans of the gels are provided in Supplementary Fig. 1.
Protein expression and purification
PfSnf2L (amino acids 250–1426) was sequence optimized for baculovirus mediated protein expression and 10×His-Tag purification. The coding sequence was cloned into the pFL plasmid and transformed into DH10Bac EM YFP cells. The bacmid DNA was isolated and transfected into Spodoptera frugiperda Sf21 cells (Invitrogen) to produce initial virus and large-scale expression50,51. Cells expressing recombinant PfSnf2L–10×His or HsSNF2L–6×His were collected and lysed in 20 mM Tris–HCl pH 7.6, 500 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 5 mM β-mercaptoethanol, 10% glycerol and 0.1% Igepal CA-630 using the Branson Sonifier 250. Purification was performed using NiNTA agarose (Qiagen) according to the manufacturer’s recommendations. The protein concentration was estimated using the Bradford assay and the purity was checked using Coomassie-stained SDS–PAGE (Extended Data Fig. 2a). The enzymes Chd4, HsSNF2L, PfSnf2Lcore, HsSNF2Lcore and PvSnf2L (Supplementary Table 4) were generated and produced as described above.
Nucleosome binding, assembly and remodelling assays
In vitro reconstituted nucleosomes in 20 mM Tris pH 7.6, 100 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol, 200 ng µl−1 BSA were incubated for 60 min at 30 °C with or without recombinant CREs. In competitive binding assays Cy3- and Cy5-labelled nucleosomes (15 nM) were incubated with increasing concentration of CREs. For non-fluorescent binding and assembly assays, 120 nM nucleosome and 700 nM CRE was used. Fluorescent remodelling assays contained 30 nM nucleosome, 1 mM ATP and variable concentrations of CREs. Assembly and remodelling reactions were stopped after 60 min (unless noted differently) by addition of 1 µg competitor plasmid DNA, and nucleosome positions were analysed by 6% native PAGE.
ATPase assays
CRE (130–700 nM) in 20 mM Tris–HCl pH 7.6, 120 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA and 10% glycerol was incubated with 500 μM ATP and 0.2 μCi 32P-γ-ATP in the absence or presence of mononucleosomes (60–350 nM) for 40 min at 30 °C. Released 32P-γ phosphate was separated from non-hydrolysed 32P-γ-ATP by thin-layer chromatography on PEI-Cellulose F plates (Merck, mobile phase: 50% acetic acid, 0.5 mM LiCl). After phosphoimaging (Typhoon FLA-9500), the signal intensities were quantified (Fuji Multi Gauge Software), the hydrolysis rate was calculated, corrected for chemical hydrolysis and normalized to CRE concentration.
Plasmid construction
A synthetic gene comprising the native nucleotides 995–1894 of PF3D7_1104200, an artificial intron with a loxP site52,53, nucleotides 1895–4278 recodonized and a 3×HA-tag-encoding sequence was ordered as a synthetic gene (IDT) and cloned into the vector pT2A-X-KO54. The resulting pT2A-Snf2L-KO plasmid contains a skip peptide sequence downstream of the PfSnf2L-coding sequence, followed by a neomycin-resistance gene to enable selection-linked integration, a second loxP site, a GFP gene and an independent human DHFR gene.
Parasite culture and transfection
P. falciparum clone 3D7 was cultured according to standard procedures in RPMI 1640 with AlbuMAX (Invitrogen), and synchronized as described previously55: schizonts were purified on a bed of 70% Percoll, incubated with new RBCs for 1–2 h, before leftover schizonts were removed with Percoll and subsequent sorbitol treatment. About 10 μg of plasmid was used for transfection of DiCre-expressing parasites using the Amaxa P3 primary cell 4D-Nucleofector X Kit L (Lonza)35,56. Successful transfection was selected with 2.5 nM of the antifolate WR99210 (Jacobus Pharmaceutical Company), starting 1 day after transfection. Resistant parasites were selected for genomic integration with 400 μg ml−1 G418 (Sigma-Aldrich). After limiting dilution of drug-resistant parasites57, genomic DNA of six clones was isolated using the Qiagen Blood and Tissue kit. Integration was confirmed by genotyping PCR using Q5 polymerase (NEB) and the primers listed in Supplementary Table 4. One stable clone was used for further phenotyping. Conditional DiCre-mediated recombination between loxP sites was performed as described previously35. For KO induction, synchronous parasites were treated with 100 nM rapamycin (Sigma-Aldrich) for 4 h (DMSO treatment as control), washed and returned to culture. Gene excision was confirmed by genotyping PCR, mRNA levels were checked in RNA-seq samples and protein levels were analysed using western blotting.
Western blot analysis
Parasites were isolated from RBCs by 0.1% saponin and protease inhibitor cocktail PIC (Roche) in PBS and boiled for 10 min in 62 mM Tris pH 6.5, 25% glycerol, 2% SDS, 0.2 M DTT, 0.05% OrangeG. Proteins were separated on 4–20% SDS gels (BioRad), and transferred onto a nitrocellulose membrane before immunoblotting. In this study, we used rat anti-HA (1:2,000, Roche) and mouse anti-Enolase (1:1,000, G. K. Jarori) antibodies, conjugated secondary antibodies and Odyssey imaging system (LiCOR Biosciences) according to manufacturer’s recommendation.
IFA
Immunofluorescence assays were performed as described previously56 with the addition of 0.0075% glutaraldehyde during fixation. Primary antibodies (rat anti-HA, rabbit anti-Gap4558, rabbit anti-Ama159) were used at a dilution of 1:500. For image acquisition, the Leica DMi8 Widefield microscope was used; image processing and quantification were performed using Fiji v.2.9.0 (ImageJ).
Analysis of parasite growth, reproduction and egress
Growth was determined by microscopy counting of parasites from Giemsa-stained thin blood films and expressed as percentage parasitaemia (percentage of infected RBCs/RBCs). For reproduction and egress assays, equal numbers of mature schizonts were Percoll-purified. For parasite reproduction, parasites were incubated in 4 ml RPMI with 1% haematocrit for 2 h, before the number of rings was quantified from Giemsa stain. Live imaging of parasite egress was performed as described previously56, and parasites were binned into groups 15 s after egress (normal/clustered merozoites). For RBC staining, RPMI was supplemented with 0.1× Phalloidin 594 Conjugate (Abcam) during live imaging. The RBC ghost location 40 s after egress was categorized (distant/attached/overlapping).
TEM analysis
Schizont-stage parasites (46 h.p.i.) were treated with compound 2 (ref. 60) for 2 h for further maturation. The parasites were then washed in PHEM buffer (2.5 mM MgCl2, 35 mM KCl, 5 mM EGTA, 10 mM HEPES, 30 mM PIPES, pH 7.2), fixed for 1 h in a solution containing 2.5% glutaraldehyde, 4% formaldehyde, 4% sucrose in 0.1 M PHEM buffer, post-fixed in 1% OsO4 plus 0.8% ferrocyanide and 5 mM CaCl2 in 0.1 M cacodylate buffer for 1 h, washed twice in cacodylate buffer (10 min, 1 h) and double-distilled water (5 min, 15 min), respectively, and dehydrated in a graded acetone series which included en bloc staining with 1% uranyl acetate in the 20% step. The cells were finally embedded in Epon812 epoxy resin to enable ultrathin sectioning. To carry out electron microscopy, the ultrathin sections were post-stained with 1% lead citrate for 2 min. Transmission electron microscopy (TEM) was performed using the JEOL F200 (JEOL) system operated at 200 kV equipped with a XAROSA 20 mega pixel CMOS camera (EMSIS).
Preparation of nuclear extracts and co‐IP experiments
Nuclei of PfSnf2L–HA and 3D7 parasites were prepared as described previously61 and treated with 0.5 U μl−1 benzonase (Sigma-Aldrich) in 20 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.65% Igepal CA-630 and PIC for 30 min at room temperature. For extraction of nuclear proteins, KCl was added to 0.4 M, incubated for 30 min at room temperature and the insoluble fraction was removed by centrifugation (5,000g, 30 min, 4 °C). The supernatant was diluted with 2.5 vol of 50 mM Tris pH 7.4, 50 mM NaCl, 1 mM EDTA, 1% Igepal CA-630 and PIC, and incubated overnight at 4 °C under constant agitation with equilibrated anti-HA magnetic beads (Thermo Fisher Scientific). Beads were pelleted using a magnetic rack, washed three times with latter buffer and three times with 50 mM Tris-HCl pH 8. The beads were dried, stored at −20 °C and used for LC–MS/MS analysis of co-immunoprecipitated proteins in PfSnf2L–HA parasites (3D7 as a negative control), in triplicates each. The collected fractions (input, flowthrough, beads) were analysed on 4–20% SDS–PAGE, silver-stained and, in an additional version, were probed with anti-HA antibodies on a western blot.
LC–MS/MS and data analysis
Beads were incubated with 10 ng μl−1 trypsin in 1 M urea and 50 mM NH4HCO3 for 30 min, washed with 50 mM NH4HCO3 and the supernatant was digested overnight in presence of 1 mM DTT. Digested peptides were alkylated and desalted before LC–MS analysis. For LC–MS/MS purposes, desalted peptides were injected into the Ultimate 3000 RSLCnano system (Thermo Fisher Scientific), separated in a 15 cm analytical column (75 μm inner diameter with ReproSil-Pur C18-AQ 2.4 μm from Dr Maisch) with a 50 min gradient from 4% to 40% acetonitrile in 0.1% formic acid. The effluent from the HPLC was directly electrosprayed into the Q Exactive HF system (Thermo Fisher Scientific) operated in data-dependent mode to automatically switch between full scan MS and MS/MS acquisition. Survey full scan MS spectra (from m/z 350 to 1,600) were acquired with resolution R = 60,000 at m/z 400 (AGC target of 3 × 106). The 10 most intense peptide ions with charge states between 2 and 5 were sequentially isolated to a target value of 1 × 105 and fragmented at 30% normalized collision energy. Typical mass spectrometric conditions were as follows: spray voltage, 1.5 kV; heated capillary temperature, 275 °C; ion-selection threshold, 33.000 counts.
MaxQuant v.1.6.14.0 was used to identify proteins and quantification was performed using iBAQ with the following parameters: database Uniprot_UP000001450_Plasmodiumfalciparum_20201007.fasta; MS tol, 10 ppm; MS/MS tol, 20 ppm Da; PSM FDR, 0.01; protein FDR, 0.01 min; peptide length, 7; variable modifications, oxidation (M), acetyl (protein N-term); fixed modifications, carbamidomethyl (C); peptides for protein quantitation, razor and unique; min. peptides, 1; min. ratio count, 2. MaxQuant iBAQ values were log2-transformed, and missing values imputed with 8. Ribosomal proteins and hits detected with only 1 peptide were excluded. Identified proteins were considered as interaction partners of the bait, if log2[Snf2L-3HA] − log2(3D7) > 3 or log2[Snf2L–3HA] − log2[3D7] > 2 and FDR < 0.05. The MS proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE62 partner repository under the dataset identifier PXD041155.
RNA‐seq and data analysis
RNA-seq analysis with or without Snf2L KO and with or without drug, respectively, was performed in triplicates. Total RNA from infected RBCs containing highly synchronized parasites was isolated using the Whole Blood Quick RNA kit (Zymo Research) according to the manufacturer’s protocol. The RNA quality was checked using 4200 TapeStation System (Agilent) and 300 ng was used as the input for Illumina Stranded mRNA Prep Ligation (Illumina). Libraries were sequenced on the Illumina NextSeq 2000 sequencing system. Sequenced reads (2 × 57 bp, paired-end, ~20 million reads per sample) were trimmed using trimmomatic (v.0.39)63 and mapped to the P. falciparum 3D7 genome v3.0 (https://PlasmoDB.org, release 52)64 using STAR (v.2.7.9a)65. PlasmoDB annotation was converted to GTF-format using gffread (v.0.12.1)66. Preprocessing and mapping quality control was done using FastQC (v.0.11.8)67, qualimap (v.2.2.2d)68, samtools (v.1.12)69 and multiqc (v.1.9)70. The pipeline was implemented with snakemake (v.5.32.0)71 and is available at GitHub (https://github.com/SimHolz/Watzlowik_et_al_2023), in addition to the R scripts. RSubread/FeatureCounts (v.2.12.2) was used to calculate read counts, while differential expression analysis was performed using DESeq272,73, with adjusted P < 0.05 used as the significance cut-off. Further analysis and visualization were done in R74 using tidyverse75 and ggpubr76. The degPatterns function from DEGreport77 was used for clustering. For expression quantile calculation, transcripts per millions normalization was used. RNA-seq data were submitted to the Gene Expression Omnibus (GEO) database (GSE228949).
RNA-seq-based cell cycle progression was estimated in R by comparing the normalized expression values of each sample to the RNA-seq data from a previous study78 using a statistical model previously described79.
PfSnf2L-KO efficacy was estimated by mapping reads to the sequence of the recodonized transfected Snf2L gene and counting reads mapped to the Snf2Lrecodon part of the gene, which is disintegrated after KO induction.
MNase‐seq and data analysis
MNase-digestion was adapted from a previous study3 with the following modifications: highly synchronous parasites were cross-linked and stopped as described. RBCs were lysed and nuclei were isolated as described for pull-down experiments. Nuclei were resuspended in 75 µl 50 mM Tris pH 7.4, 4 mM MgCl2, 1 mM CaCl2, 0.075% NP40, 1 mM DTT, PIC with 0.75 U Micrococcal nuclease (Worthington Biochemicals) and 50 U exonuclease III (NEB). Each sample was aliquoted in 3 × 25 µl and incubated for 2.5, 5 and 10 min at 37 °C under agitation (low, mid and high digestion), before the reaction was stopped by adding 25 µl 2% Triton X-100, 0.6% SDS, 300 mM NaCl, 6 mM EDTA/PIC and placed at 4 °C. De-cross-linking was performed at 45 °C overnight after adjusting to 1% SDS, 0.1 M NaHCO3 and 0.5 M NaCl. Proteins were digested by addition of 40 µg of proteinase K (Zymo Research) and incubation for 1 h at 55 °C. Subsequently, DNA was isolated using the EXTRACTME DNA clean-up kit and Micro Spin columns (Blirt) according to the manufacturer’s recommends. Sequencing and data analysis is described in Supplementary Note 1.
ADP biosensor assay
ATPase activity and inhibition were measured using the ADP biosensor assay: in 20 µl, 0.2 µM TMR-maleimide-labelled ParM (prepared as described previously80) was mixed with 125 µM ATP, 100 ng plasmid pT11 DNA, 5 µM H4 peptide (amino acids 8–25, AnaSpec) in 10 mM Tris pH 8.6, 1.5 mM MgCl2, 100 mM KCl, 0.01% pluronic in presence (or absence as a negative control) of 0.4 µM recombinantly expressed PfSnf2L and NH125 at varying concentrations. The resulting ADP binding to ParM is expressed by increasing fluorescence intensity and was kinetically measured at 28 °C in 2 min intervals over 2 h in the Tecan infinite F500 reader. The signal was normalized to timepoint 0.
Toxicity assay for Plasmodium, Toxoplasma and HeLa
Toxicity for Plasmodium was determined by culturing in presence of NH125 and DMSO for 72 h and subsequent quantification of parasitaemia in Giemsa-stained blood films. Toxicity for Toxoplasma gondii was tested using a plaque assay, whereby 1,000 T. gondii tachyzoites inoculated onto human foreskin fibroblasts as described previously81 were treated with NH125 and DMSO, fixed and stained 6 days later and evaluated for plaque formation. Toxicity on human HeLa cells, cultured as described previously49, was investigated by 48 h NH125/DMSO treatment and subsequent monitoring of metabolic activity using the Cell Proliferation Kit II-XTT (Sigma-Aldrich) according to the manufacturer’s recommendations. Half-maximal effective concentration (EC50) values were obtained by fitting the dose–response model using three-parameter log-logistic models and estimated for T. gondii.
Gametocyte induction
Sexual commitment was induced using the nutrient deprivation induction method as described previously82. In brief, parasites were tightly synchronized using Percoll (63% isotonic solution) density-gradient centrifugation to isolate mature-stage schizonts and allow invasion of naive RBCs, followed by treatment with 5% sorbitol to kill the remaining schizonts in the culture and retain only young rings (day 0). The assay was performed in six-well plates and started at 1.5% parasitaemia and 3% haematocrit. For a high-throughput sexual-commitment–conversion assay in a 96-well format, parasites were plated at 0.5% parasitaemia and 2.5% haematocrit. Drug treatment with NH125 and/or rapamycin treatment for the inducible KO line were started at 10 h.p.i. in complete medium supplemented with choline chloride as indicated in Extended Data Fig. 9a. To induce sexual commitment, parasites were shifted to minimal fatty acid (mFA) medium at 20–24 h.p.i. (day 1). mFA medium was prepared by supplementing incomplete medium (RPMI-1640 without AlbuMAX) with 0.39% fatty-acid-free BSA (Sigma-Aldrich), 30 µM oleic acid (Sigma-Aldrich) and 30 µM palmitic acid (Sigma-Aldrich). Then, 22–26 h after induction (day 2), the mFA medium was replaced with complete RPMI medium (with AlbuMAX). At this timepoint, in the high-throughput assay, the parasites were treated with 50 µM E64 to block merozoite egress and measure the sexual commitment rate based on the number of AP2–GGFP-positive schizonts. On day 3, parasitaemia was quantified using Giemsa-stained blood smears or by flow cytometry using Hoechst staining, and 20 U ml−1 heparin was added to the culture until day 6. The medium was changed daily until quantification of gametocytemia on day 9 by Giemsa staining to calculate sexual conversion rate as follows: sexual conversion rate = gametocytemia on day 9/parasitaemia on day 3. Within the sexual-commitment-conversion assay, parasite conversion was analysed with flow cytometry on day 6 after staining with TubulinTracker Deep Red and Hoechst, or on day 7 based on GEXP-02 positive cells (Extended Data Fig. 10).
Dose–response assays
Half-maximal inhibitory concentrations (IC50 values) for NH125 were determined against various strains by exposing ring-stage parasites to a range of drug concentrations (twofold serial dilutions starting from 5 µM) in 96-well plates. Assays were seeded at 0.2% starting parasitaemia and 1% haematocrit. Parasite growth in each well was determined after 72 h by flow cytometry after staining with Mito Tracker Deep Red and Hoechst. Assays were performed in duplicate. IC50 values were calculated from three to four independent replicates by nonlinear regression analysis in GraphPad Prism.
ChIP–seq and data analysis
ChIP experiments were performed on PfSnf2L–HA parasites at 10, 22, 34 and 46 h.p.i. in duplicate with an additional no-epitope control on WT 3D7 parasites, respectively. Highly synchronous parasites were cross-linked and stopped as described previously3. RBCs were lysed and nuclei were isolated as for co-IP experiments. Nucleus isolation was followed by chromatin sonication using the Covaris Focus-Ultrasonicator (5% duty cycle, 75 W peak incident power, 200 cycles per burst, 7 °C, 5 min), protein–chromatin complex immunoprecipitation using anti-HA antibodies and DNA purification (Qiagen MinElute Kit) as performed previously83,84. All ChIP libraries (with paired non-immunoprecipitated input control samples) were prepared as described previously83,84, and checked for library sample quality (high-sensitivity DNA Qubit Fluorometer) and sequence length (Agilent TapeStation 4150) before sequencing on the Illumina NextSeq 2000 system for P1 150 × 150 paired-end sequencing. ChIP reads were first trimmed using Trimmomatic v.0.32.3 (<30 phred, SLIDINGWINDOW:4:30 option)63. The pre- and post-trimming read quality was assessed using FastQC (v.0.11.9)67. Filtered and trimmed reads were then mapped using BWA-MEM (v.0.7.17.2)85 using paired-end simple Illumina mode to the P. falciparum 3D7 genome (https://PlasmoDB.org; release 52) and filtered for multi-mapped reads (MAPQ = 1 option). Enrichment was calculated as log2[ChIP/input] using deepTools bamCompare (v3.5.2)86, averaged for replicates and aligned to +1 nucleosome as for nucleosome occupancy. ChIP–seq data were submitted to the GEO (GSE237217).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.