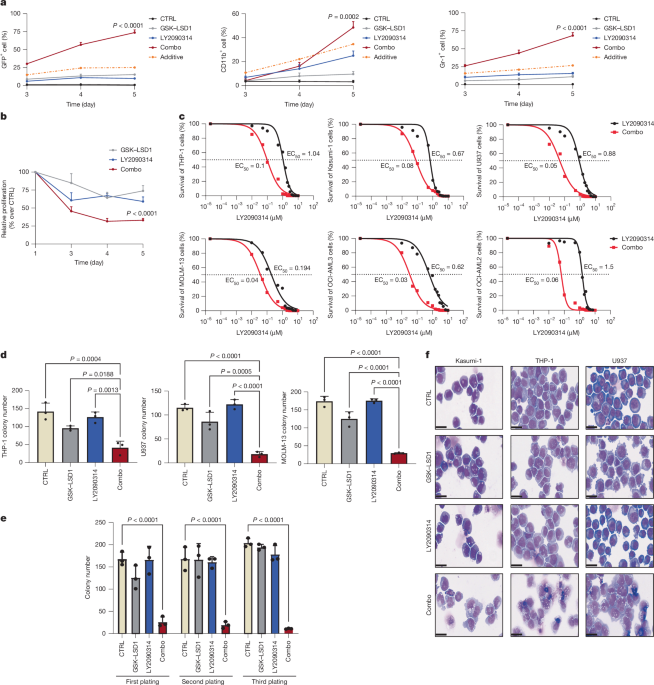
Small-molecule inhibitor screen
ER-HOXA9 cells were prepared in media with 50 nM GSK–LSD1 (Selleck Chemical) and seeded at a density of 50,000 cells per millilitre in 200 µl volume per well of a flat-bottom 96-well plastic plate (Genesee Scientific) using a Combi Reagent Dispenser (Thermo Fisher). Drugs in 100% DMSO (Sigma-Aldrich) were pin transferred (V&P Scientific) from 384-well stock plates into the 96-well plates containing our cells at approximately 300 nl drug stock per well. Cells treated with GSK–LSD1 alone served as negative control, whereas cells treated with GSK–LSD1 and 100 nM cytarabine (Sigma-Aldrich), a known synergistic combination, served as a positive control. Plates were incubated for 5 days and analysed on an iQue Screener Plus-VBR flow cytometer (Intellicyt) running the Forecyt acquisition and analysis software (v9.0). Monocytic differentiation was assessed using an internal Lyz2–GFP marker (blue laser channel at 488-nm excitation and 530-nm emission). Viability was calculated by dividing the number of live cells by the number of total cells, and differentiation was calculated by dividing the number of Lyz2–GFP+ cells by the number of live cells.
Cell culture
ER-HOXA9 cells were grown in RPMI-1640 medium supplemented with 10% of fetal bovine serum (FBS), 100 ng ml−1 stem cell factor (SCF; 78064, Stemcell Technologies), 4 mM glutamine, 1% penicillin–streptomycin and 0.5 mM β-oestradiol (E2; E4389, Sigma-Aldrich). HOXA9–MEIS1 cells were similarly passaged as ER-HOXA9 cells except without E2. RN2 cells were cultured in RPMI-1640 medium supplemented with 10% FBS, 20 mM l-glutamine, 10 mM sodium pyruvate, 10 mM HEPES (pH 7.3), 1% penicillin–streptomycin and 50 μM β-mercaptoethanol.
THP-1 and U937 were grown in RPMI-1640 medium supplemented with 10% of FBS, 4 mM glutamine and 1% penicillin–streptomycin. MOLM-13 and Kasumi-1 were grown in RPMI-1640 medium supplemented with 20% of FBS, 4 mM glutamine and 1% penicillin–streptomycin. OCI-AML2 and OCI-AML3 were grown in α-MEM (with ribonucleosides and deoxyribonucleosides) with 20% FBS, 4 mM glutamine and 1% penicillin–streptomycin. Cells were maintained at 37 °C and 5% CO2 with routine testing to confirm lack of mycoplasma infection.
Drug combination assay and synergy score analysis
The drug synergy assay was performed as previously described69. In brief, cells were seeded into 96-well plates and exposed to various concentrations of inhibitors, both individually and in combination. Cell viability was quantified using Cell-Titer-Glo (Promega) and normalized to DMSO to calculate the inhibitory response. The resulting data were analysed using the SynergyFinder web application (https://synergyfinder.fimm.fi), which generated dose–response matrices for each drug combination. Synergy scores were calculated using the highest single-agent model to assess drug interactions. Heatmaps were generated to visualize the results, with the following interpretation thresholds: synergy scores below −10 indicated antagonism, scores between −10 and 10 suggested an additive effect, and scores above 10 indicated synergy.
Human primary AML samples
Bone marrow or peripheral blood samples were collected after informed consent from patients with AML using protocols approved by an Institutional Review Board at the Helsinki University Hospital (permit numbers 239/13/03/00/2010 and 303/13/03/01/2011) in compliance with the Declaration of Helsinki. Mononuclear cells were isolated from bone marrow or peripheral blood samples by Ficoll-Paque Premium (GE Healthcare) density gradient separation and viably frozen and stored in liquid nitrogen before further analyses.
LSK cell sorting and colony formation
Eight-week-old C57BL/6J mice were euthanized, and bone marrow was harvested from the femurs and tibias of both legs. Haematopoietic progenitor cells were enriched using the EasySep Mouse Hematopoietic Progenitor Cell Isolation Kit (19856, StemCell Technologies). Haematopoietic progenitor-enriched cells were then stained with antibodies to mouse KIT (clone: ACK2, 567818, BD Bioscience), Sca-1 (clone: E13-161.7, 753334, BD Bioscience), lineage markers (CD3, CD11b, CD19, B220, Gr1 and Ter119; 155606, 101212, 115512, 103212, 108412 and 116212, respectively, BioLegend), and Fixable Viability Stain 575V (565694, BD Bioscience). LSK cells were sorted using a FACS Aria Fusion (BD Bioscience). For the colony formation assay, 2,000 LSK cells were resuspended in 100 μl of IMDM and added to 1.5 ml of methylcellulose media (M3434, StemCell Technologies). Colonies were counted after 10 days of incubation at 37 °C. ImageJ (v1.54g) was used for the analysis of colonies morphology images.
Ex vivo drug sensitivity testing of primary AML cells
GSK–LSD1 and LY2090314 (MedChem Express) were dissolved in 100% DMSO and dispensed on Nunc 96-well polystyrene V-bottom plates (Thermo Fisher Scientific) using the Echo 550 Acoustic Dispenser (Labcyte) in seven different concentrations. GSK–LSD1 was plated in a concentration range of 1–250 nM and LY2090314 in a range of 50–500 nM as single agents and in combination. 0.1% DMSO was used as a negative control, and 100 µM benzethonium chloride (Sigma-Aldrich) was used as a positive control for total cell death.
Frozen mononuclear cells were thawed and suspended in 12.5% conditioned medium composed of RPMI-1640 medium (Corning) supplemented with 12.5% HS-5 cell-derived conditioned medium, 10% FBS 2 mM l-glutamine and penicillin–streptomycin (100 U ml−1)70, and then treated with DENARASE (250 U µl−1, c-LEcta) to degrade DNA released from dead cells; the cells were left to recover for 4 h in 12.5% conditioned medium. The cells were plated onto pre-drugged plates at a density of 50,000 cells per well and incubated with the drugs for 5 days (at 37 °C at 5% CO2). After incubation, the cells were centrifuged (at 500g for 5 min) and resuspended in staining buffer (RPMI-1640, 10% FBS, 2 mM l-glutamine and 100 U ml−1 penicillin–streptomycin). The cells were stained with antibodies to CD45–FITC (BD Pharmingen), CD34–APC (BD Pharmingen), CD15–PE–Cy7 (BioLegend), CD14–BV421 (BD Biosciences) and CD11b–BV605 (BD Horizon) for 30 min at room temperature in the dark. Subsequently, the cells were centrifuged (at 500g for 5 min) and excess antibodies were removed. The cells were resuspended and stained with PE annexin V and seven-amino actinomycin D in annexin V-binding buffer (BD Pharmingen) for 15 min at room temperature in the dark. The cells were analysed using the iQue Screener Plus-VBR flow cytometer, and gating was done with ForeCyt software (version 9.0, Intellicyt). Data were processed and analysed using R software (v4.2).
Seeding of organoids with primary patient samples and drug treatment
Human bone marrow organoids were generated from a fluorescent human induced pluripotent stem (iPS) cell line as previously described52,53. The fluorescent iPS cell line, MCND-TENS2-mScarlet3, was obtained through CRISPR–Cas9-mediated knock-in of mScarlet3 at the AAVS1 safe harbour locus in the parental line MCND-TENS2 (registered at https://hpscreg.eu/cell-line/RTIBDi001-A), performed by the iPS Cell Core Facility at the US National Institutes of Health (NIH). On day 14 of organoid differentiation, individual mScarlet+ organoids were seeded into 96-well ultra-low attachment plates. Cryopreserved mononuclear cells from samples from patients with AML were then engrafted into the organoids at a density of 10,000 cells per organoid, with 8 organoids for each treatment condition. The organoids were subsequently cultured in StemPro-34 SFM medium (10639011, Thermo Fisher Scientific) supplemented with 2% KnockOut serum (10828028, Thermo Fisher Scientific), 2% chemically defined lipids (11905031, Thermo Fisher Scientific), 0.5% penicillin–streptomycin and cytokines (10 ng ml−1 of SCF, FLT3-L, TPO, IL-6, G-CSF and GM-CSF, and 5 ng ml−1 of IL-3). Twenty-four hours post-engraftment, the engrafted organoids were treated with either vehicle control, 25 nM GSK–LSD1, 50 nM LY2090314 or a combination of inhibitors for 5 days. Following treatment, the eight organoids from the same condition were pooled to minimize variations, and they were then dissociated using collagenase D (11088866001, Roche)71 and analysed by flow cytometry to determine the percentage of mScarlet−CD11b+ cells.
Phosphoflow analysis
After thawing and DENARASE treatment as previously described, mononuclear cells from samples from patients with AML were plated onto pre-drugged Nunc 96-well V-bottom plate at a density of 200,000 cells per well and incubated with 50 nM GSK–LSD1, 100 nM LY2090314 and the combination of both drugs for 5 days (at 37 °C at 5% CO2). After incubation with the drugs, the cells were washed with PBS, centrifuged (at 1,000g for 4 min) and stained with Zombie Yellow (BioLegend) viability marker for 30 min in the dark at room temperature. The cells were washed with staining buffer (5% FBS in Dulbecco’s phosphate-buffered saline (DPBS)) and stained with surface markers for CD45–BV786 (BD Biosciences), CD38–BV421 (BD Biosciences), CD34–APC–Cy7 (BioLegend) and CD11b–BV605 (BD Horizon) for 30 min at room temperature. The cells were fixed in 1.5% paraformaldehyde solution in PBS pre-warmed to 37 °C for 15 min at room temperature. Fixed cells were centrifuged (at 1,000g for 4 min), washed with staining buffer and centrifuged again with the same settings. The cells were resuspended in ice-cold methanol and incubated at 4 °C for 30 min, after which the cells were washed twice with staining buffer with centrifugation (at 1,000g for 4 min). The cells were stained with β-catenin–AF488 (BD Pharmingen) for 1 h at room temperature. After incubation, the cells were washed with staining buffer, centrifuged (at 1,000g for 4 min), then resuspended and analysed on an iQue PLUS flow cytometer, and data were analysed using the Forecyt software (v9.0). Data were processed and analysed using R software (v4.2).
In vivo study
HOXA9–MEIS1-overexpressing leukaemia cells previously developed by Sykes et al.24 were virally transduced to express Luciferase and GFP for in vivo tracking. Cells expressing GFP were twice sorted and used to establish syngeneic mouse models of AML in 6–8-week-old female C57BL/6J mice purchased from Jackson Laboratory. Animals were maintained at Boston Children Hospital’s ARCH facility and treated according to all protocols approved by the Institutional Animal Care and Use Committee under protocol number 16-09-3230R. The mice received sublethal radiation of 350 cGy 16–20 h before tail-vein injection of 0.5 × 104 leukaemia cells in 100 µl PBS to establish a measurable residual disease model of AML. Leukaemia engraftment and therapy response were monitored using whole-body IVIS imaging through retro-orbital injection of luciferin. Mice were randomized into four treatment groups after engraftment was observed 7 days post-injection.
NSG (NOD.Cg-PrkdcSCID Il2rgtm1Wjl/SzJ) mice were purchased from Charles River and irradiated with a sublethal dose of 1.5 Gy 24 h before intravenous injection. For the OCI-AML3 model, approximately 1 million cells were transplanted via tail-vein injection into 6–8-week-old male or female NSG recipient mice. To assess leukaemia development, peripheral blood was collected from the mice, stained for human CD45 and analysed by flow cytometry; treatment was initiated 13 days after transplantation, once hCD45+ cells were detected in the peripheral blood. For luciferase-expressing PDX samples, AML-372 (DNMT3A-WT) and AML-579 (DNMT3A-mutant) models50,51, about 700,000 and 1 million cells, respectively, were transplanted via tail-vein injection into 6–8-week-old female NSG recipient mice and treatment was initiated 13 days after transplantation. Engraftment and leukaemia burden were evaluated using a bioluminescence imaging system following the intravenous administration of d-luciferin (P1043, Promega). For in vivo treatments, either GSK–LSD1 (0.25 mg kg−1), LY2090314 (10 mg kg−1) or a combination of both was administered via intraperitoneal injections every alternate day for 1 week (HOXA9–MEIS1 model) or 2 weeks (OCI-AML3 model and PDX models). Animals were monitored daily, and body weights were measured throughout the treatment period. Mice exhibiting signs of distress, rough fur, hunchback and reduced motility were euthanized by a schedule 1 method. Kaplan–Meier survival curves were generated using GraphPad Prism (v10) software. All cages were on a 12-h–12-h light–dark cycle (lights on at 07:00) in a temperature-controlled and humidity-controlled room. Room temperature was maintained at 19–23 °C, and room humidity was maintained at 45–65%. All mouse procedures were carried out in accordance with UK Animals (Scientific Procedures) Act 1986 and University of Oxford Animal Welfare and Ethical Review Body approval under Project license (PPL) number PP4128654.
Dot blots
Purified total RNA from treated cells were subjected to digestion with mock, RNase T1 (AM2283, Thermo Fisher Scientific) and RNase III (AM2290, Thermo Fisher Scientific) in their respective buffers and according to the manufacturer’s instructions, or RNase A (EN0531, Thermo Fisher Scientific) under high-salt condition (350 mM NaCl). The digestion was deactivated by the addition of TRIzol and RNA samples extracted using the TRIzol manufacturer’s protocol (R2053, Zymo Research). Equal volumes (3 µl) of purified RNA were dotted on Hybond N+ membrane (RPN119B, GE Healthcare), air dried for 10–15 min at room temperature, then UV crosslinked in a UV stratalinker 2400 (Stratagene) two times. The membrane was blocked for 1 h in blocking buffer (5% milk diluted in 0.01% PBS-T) and probed with J2 antibody (RNT-SCI-10010500, Jena Bioscience) rocking overnight at 4 °C. On the next day, the membrane was washed three times in PBS-T, rocking for 10 min at room temperature per wash and probed with secondary goat-anti-mouse horseradish peroxidase (HRP) antibody in 5% milk at room temperature for 1 h. Membrane was washed three times in PBS-T, rocking for 10 min at room temperature per wash, and enhanced chemiluminescence (ECL) was applied for chemiluminescent development. To detect total nucleic acid loading, the membrane was then incubated in 0.5% methylene blue in 30% EtOH to visualize the presence of RNA.
CRISPR–Cas9 gene knockouts
CRISPR gene editing was performed using the Integrated DNA Technologies (IDT) Alt-R CRISPR–Cas9 System as per the manufacturer’s protocol. In brief, Alt-R CRISPR–Cas9 CRISPR RNA (crRNA) was mixed with Alt-R CRISPR–Cas9 trans-activating crRNA (tracrRNA) and Alt-R HiFi S.p. Cas9 Nuclease V3 to assemble the ribonucleoprotein complex. Subsequently, this complex was electroporated into target cells using the Neon transfection system, using a pulse voltage of 1,400, a width of 10 ms and three pulses. Alt-R CRISPR–Cas9 negative control crRNA #2 was used for the creation of non-targeted controls. Specific gene knockouts were generated using guide RNAs listed in Supplementary Table 13 that were selected using the IDT predesign and selection tool.
Gene knockdown by shRNA
Target sequences for shRNA knockdown of the genes encoding GSK3α and GSK3β were sourced from existing literature36 and oligos were ordered from IDT. In brief, shRNA oligos were annealed and ligated into the pLKO.1-Puro (Addgene #10878) plasmid backbone (digested with AgeI and EcoRI) overnight and subsequently transformed into NEB stable-competent Escherichia coli (C3040H, NEB). Colonies were screened for correct insertion using primers flanking approximately 100 bp upstream and downstream of the AgeI and EcoRI restriction sites. Positive clones were isolated and sent for sequencing before co-transfection with pCMV-dR8.2 and pCMV-VSVG into HEK293T cells for lentivirus production.
Target sequences used for shRNA knockdown experiments listed in Supplementary Table 14.
Total RNA extraction and RT–PCR
Total RNA isolation and DNaseI treatment was performed using the Direct-zol RNA MiniPrep kit (R2053, Zymo Research) as per the manufacturer’s protocol. Reverse transcription of 1 μg of RNA per sample was performed using SuperScript IV Vilo (11756050, Thermo Fisher Scientific) as per the manufacturer’s protocol and quantified by spectrophotometer (ND1000 NanoDrop). From 5 ng to 10 ng of cDNA was used to perform quantitative PCR (qPCR) using SYBR Select Master Mix (4472908, Thermo Fisher Scientific). All the qPCR amplifications were performed in the Step One Plus system (Applied Biosystems).
Gene expression values were calculated by the ΔCq method, using GAPDH as the housekeeping gene, and resulting experimental target values were normalized to the global mean of the control group. Normalized fold change was plotted using GraphPad Prism software. The sequences of the primers used in this study are listed in Supplementary Table 15.
In vitro studies and viability assays
Approximately 2,500 cells were plated in triplicates in 96-well plates for 5 days. For in vitro experiments, cells were treated with 50 nM GSK–LSD1 (SML1072, Sigma-Aldrich), 50 nM bomedemstat (IMG-7289; HY-109169B, MedChem Express), 500 nM TAK-418 (HY-138830, MedChem Express), 100 nM LY2090314 (HY-16294, MedChem Express) and 100 nM elraglusib (9-ING-41; HY-113914, MedChem Express).
Cell viability was determined using a Cell Titer-Glo luminescent cell viability assay (G7572, Promega). Data were presented as proliferation present by comparing the treated groups with the vehicle-treated cells.
Colony-forming unit assay
Leukaemia cell lines
Approximately 1,000 cells (for human leukaemia cell lines) and 500 cells (for mouse leukaemia cell lines) were initially plated in triplicates in the methylcellulose medium (MethoCult GF, H4435, StemCell Technologies) pre-added with vehicle, GSK-LSD1, LY2090314 or a combination of inhibitors. For serial replating, cells isolated from colonies in the previous plating were seeded again in the same semi-solid medium. Colony-forming units were scored every 7–10 days post-seeding.
Human CD34+ umbilical cord blood cells
Human cord blood CD34+ cells (200-0000, StemCell Technologies) were plated in methylcellulose (MethoCult H4534 Classic, StemCell Technologies). For each condition 5,000 cells were plated in 35-mm dishes in the presence of inhibitors. After 14 days, haematopoietic colonies were scored.
Human primary AML samples
Patient samples were thawed and cultured in StemSpan SFEM II (StemCell Technologies) supplemented with human recombinant Flt3/Flk-2, human recombinant IL-3, human recombinant GM-CSF, human recombinant IL-6, human stem cell factor and human recombinant G-CSF (StemCell Technologies) for 24 h. The cells were then treated with DMSO or inhibitors in methylcellulose medium (MethoCult H4535 or MethoCult H4534 Classic, StemCell Technologies) plated at 25,000 cells per millilitre on 35-mm culture dish and cultured for 7–10 days to form colonies.
Wright–Giemsa staining
The cells collected from culture plates were spun onto a cytological slide by using a cytospin centrifuge (Cytospin 4 Cytocentrifuge). Then, slides were stained using the May–Grünwald–Giemsa staining method. The fixed cells were stained for 8 min in May–Grünwald stain (MG500, Sigma-Aldrich), then slides were sequentially washed 6 times in deionized water and then incubated for 30 min with Giemsa stain (1092041000, Sigma-Aldrich) and diluted with 19 volumes of distilled water. After this step, the cytological slides were rinsed again three times in distilled water and air dried. For long-time storage, a coverslip was attached to the slides by Eukitt mounting medium, which is an adhesive and specimen preservative that can be used manually and in automated coverslipping equipment. The slides were scanned using the NanoZoomer S210 slide scanner.
TCF/LEF luciferase reporter assay
HCT116 and THP-1 cells were plated at 100,000 cells per well in 12-well plates, and following a 24-h incubation were infected with 30 µl TCF/LEF luciferase reporter lentivirus (BPS Biosciences). Following 48 h, cells were replated and selected for 3 days using puromycin. Then, cells treated with inhibitors in the presence and absence of WNT3A (40 ng ml−1) for 5 days. TCF/LEF activity was assessed using ONE-Step Luciferase reagent per recommended protocol (BPS Biosciences).
RNA-seq protocol and analysis
For freshly cultured cells, total RNA isolation and DNaseI treatment were performed using the Direct-zol (TM) RNA MiniPrep kit (R2053, Zymo Research) as per the manufacturer’s protocol. Library preparation was conducted using the NEBNext UltraII RNA library kit (E7770S/L, NEB). Sequencing was conducted on an Illumina NextSeq 2000.
For ER-HOXA9 cell RNA-seq analysis, Fastq reads were aligned to the mouse genome (mm10) using STAR 2.7.0f. Read counts were mapped to genes using featureCounts. Differential analysis of gene expression was performed using DESeq2 (1.34.0). Only genes with more than 10 total counts when summed across all samples were considered. For THP-1 RNA-seq analysis, RNA-seq analysis was conducted using the EdgeR (3.50.3) limma (3.36.0) workflow. Reads were quantified using featureCounts, creating the raw gene count matrix. Data-quality metrics were investigated, and the limma voom normalization was applied to obtain counts per million (CPM) normalization and trimmed means of M values and normalization to finalize the differential expression analysis. The normalization accounted for sequencing depth. The log-CPM values were calculated, and adjusted P < 0.01 were considered significant. Genes responding synergistically to combo treatment were defined as those with adjusted P < 0.01 and fold change > 3 in combo versus vehicle and were also not significant at adjusted P < 0.01 and fold change > 1.5 in either GSK–LSD1 or LY2090314 versus vehicle. In THP-1 cells, genes responding synergistically to combo treatment were defined as those with adjusted P < 0.01 and more than 3× change in combo versus vehicle and not significant at P < 0.05 (non-adjusted) in either GSK–LSD1 or LY2090314 versus vehicle in THP-1 cells. Heatmaps of synergy genes and other gene lists were generated using Heatmapper.ca using Pearson correlation and average linkage settings. For pathway-level analysis, gene lists were either submitted to EnrichR72,73,74 or GSEA75,76 (4.3.3) was used. For GSEA, CPM-normalized data were used as inputs and GSEA MSigDB (2024.1) gene set compendia, or manually curated gene sets were used for enrichment using genes for permutations and default settings. Manually curated gene sets not in the MSigDB database included the Sykes terminal differentiation gene sets24, the LSC47 leukaemia stem cell signature77 and the Somervaille leukemia stem cell signatures78.
ATAC-seq protocol and analysis
ATAC-seq was performed as previously described23. In brief, cells were treated with DNAseI (EN0521, Life Tech) to remove genomic DNA contamination. Live cell samples were quantified and assessed for viability and after cell lysis and cytosol removal, nuclei were treated with Tn5 enzyme (20034197, Illumina) for 30 min at 37 °C and purified with the Minelute PCR Purification Kit (28004, Qiagen) to produce tagmented DNA samples. Tagmented DNA was barcoded with Nextera Index Kit v2 (FC-131-2001, Illumina) and amplified via PCR before an SPRI Bead cleanup to yield purified DNA libraries. Sequencing was performed on an Illumina HiSeq instrument (4000 or equivalent). Fastq files were subjected to quality control with FastQC (0.11.9) and then trimmed with Cutadapt (2.1) with reads less than 20 nucleotides being filtered out. Reads were then mapped against mm10 with Bowtie2 (2.4.4), and duplicate reads were removed with samtools (1.15.1) rmdup, and bam files were converted to bed files with bedtools (2.30.0) bamtobed. Peaks were then called with MACS2 (2.2.7.1) with replicates being merged for downstream analyses. For heatmaps and PCAs, matrices were generated with deeptools (3.5.1) computeMatrix, and heatmaps and PCAs were generated with deeptools plotHeatmap and ggplot2 (3.4.2), respectively. IFNα signal profiles were generated with deeptools plotHeatmap using the IFNA promoter regions as an input. IFNA promoters were extracted using the ChIPseeker (1.42.0) R (4.3.0) package. ATAC-seq tracks were visualized with Integrative Genomics Viewer (2.16.2). Homer (5.1) was used for motif enrichment analyses using default settings. All operations were performed using default settings unless otherwise noted.
CUT&RUN protocol and analysis
In brief, 250,000 cells were washed in 1 ml of wash buffer (20 mM HEPES pH 7.5, 0.5 mM spermidine and 120 mM NaCl) and centrifuged at 600g at room temperature three times, and the pellet were resuspended in 100 μl of wash buffer per reaction. Following this, 10 μl activated concavalin A beads per reaction was added, and the mixture was incubated at room temperature for 10 min with intermittent shaking. Captured cells were resuspended in 50 μl of antibody-binding buffer (wash buffer with digitonin 0.05% and EDTA). Primary antibodies, including negative and positive controls, were added, and the tubes were nutated overnight at 4 °C. On the following day, the samples were washed twice in dig wash buffer (wash buffer with 0.05% digitonin), and a master mix of pAG-MNase was prepared and added to each sample. After nutating at 4 °C for 1 h, the samples were washed to remove unbound pAG-MNase. Tubes were cooled to 0 °C for 5 min, then supplemented with CaCl2 to promote MNase digestion at 4 °C for 2 h followed by the addition of 2X STOP buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA and 0.05% digitonin) and incubation at 37 °C for 10–15 min. Following a brief spin and magnetic separation, the supernatant containing enriched target-bound chromatin was proceeded directly with DNA cleanup with the Zymo DNA Clean&Concentrator-5 kit (Zymo Research). DNA libraries were prepared by the NEBNext Ultra II DNA library kit (E7645S) as per the manufacturer’s instructions.
CUT&RUN samples were processed via Nextflow (21.10.6), using the nf-core CUT&RUN pipeline (v3.0.0)79. Samples were aligned to the hg38 reference genome. Adapters were trimmed using Trim Galore (0.6.6), and paired-end alignment was performed using Bowtie2 (2.4.4). Mapping rates, GC content and other sample quality metrics were derived from nf-core via MultiQC. Peak calling was finalized using SEACR80 (1.3) with a standard peak threshold of 0.05 and spike-in calibration performed with the Saccharomyces cerevisiae genome. Heatmap and PCA analyses by gene and peak were performed using deepTools (3.5.1). Downstream peak-based analyses were done with peak bed files from replicate experiments being merged. Merging was done using bedTools (2.30.0) concatenate to combine peak files, bedTools sort to order peaks, and bedTools merge to merge peak regions. Motif enrichment analysis, track visualization and signal over gene set promoter regions (IFNα and Sykes myeloid differentiation top 200 gene promoter regions) were done as described in the ATAC-seq analysis section in the Methods. Genomic distribution analyses were done with the ChIPseeker (1.42.0) R (4.3.0) package, and peak and gene overlaps were quantified with bedTools intersect. The sequences of the primers used for CUT&RUN-qPCR are listed in Supplementary Table 16.
Clinical dataset survival analysis
Patient data used in this study were taken from the cohort used in Bottomly et al.81 and referred to as the OHSU patient dataset. For signature score analyses, patients were scored according to their match in expression to a list of signature genes (upregulated genes only, downregulated genes only or both upregulated and downregulated genes) using the singscore (1.26.0) R (4.3.0) package82. If upregulated signature genes only were used, the patient score is high if the patient upregulates those genes. If downregulated signature genes only were used, the patient score is high if the patient downregulates those genes. If both upregulated and downregulated signature genes were used, the upregulated gene score and downregulated gene scores are combined. Pearson correlation was used to quantify correlations between signature scores among the patients. Components of signatures used for score correlations in Fig. 5j were: upregulated genes of the ER-HOXA9 synergy signature and downregulated genes of the Somervaille LSC signature (upper left); downregulated genes of the ER-HOXA9 synergy signature and total score of the MSigDB Wnt Signaling signature (upper right); upregulated and downregulated genes of the ER-HOXA9 synergy signature and upregulated genes of the MSigDB Hallmark Interferon Alpha Response signature (lower left); and upregulated genes of the MSigDB Hallmark Interferon Alpha Response signature and downregulated genes of the Somervaille LSC signature (lower right). For survival analyses, Kaplan–Meier plotting was performed using the ggsurvplot function of the survminer R (4.3.0) package. Patients were stratified by median synergy score enrichment using the downregulated genes of the ER-HOXA9 synergy signature. Statistical significance of survival data was tested with log-rank tests using the survdiff function of the Survival (3.8-3) R (4.3.0) package.
Immunoblotting
Cells were lysed in a lysis buffer (0.1% SDS, 400 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl and 1% Triton) plus protease inhibitor cocktail (11836170001, Sigma-Aldrich). Protein quantification was performed using a BCA assay (Promega). Of proteins, 20–40 μg was mixed with Laemmli (Bio-Rad) and denatured for 10 min at 95 °C. Cell lysates were loaded onto each lane of a 4–20% Mini-PROTEAN TGX Gel (Bio-Rad). The proteins were then transferred to a Trans-Blot Turbo Midi Nitrocellulose Transfer membrane (Bio-Rad). Following this, the membrane was blocked using 5% BSA and incubated with primary antibodies overnight at 4 °C. The next day, after three washes with 1% TBS-T (each wash 10 min), membrane was incubated with the proper secondary HRP antibodies, diluted in 5% BSA, for 30–60 min at room temperature. The membrane was washed again with 1% TBS-T three times, and ECL was applied for membrane development. The Bio-Rad ChemiDoc was used for the acquisition of western blot images. Antibodies included: β-catenin (D10A8) (dilution 1:1,000; 8480S, Cell Signaling), IRF-7 antibody (F-1; dilution 1:1,000; sc-74471, Santa Cruz Biotechnology), α-tubulin (DM1A; dilution 1:1,000; sc-32293, Santa Cruz Biotechnology), vinculin (42H89L44; dilution 1:1,000; 700062 Thermo Fisher Scientific), β-actin (13E5; dilution 1:1,000; 4970S, Cell Signaling), GSK3α (dilution 1:1,000; 9338, Cell Signaling), GSK3β (D5C5Z; dilution 1:1,000; 12456, Cell Signaling), anti-GSK3α and anti-GSK3β (phospho-Y216 + Y279) antibody (M132; dilution 1:1,000; ab45383, Abcam), STAT1 (dilution 1:1,000; 9172, Cell Signaling), phospho-STAT1 (Tyr701) monoclonal antibody (ST1P-11A5; dilution 1:1,000; 33-3400, Thermo Fisher Scientific), anti-rabbit IgG, HRP-linked antibody (dilution 1:5,000; 7074S, Cell Signaling) and anti-mouse IgG, HRP-linked antibody (dilution 1:5,000; 7076S, Cell Signaling).
Immunoprecipitation
Of cleared protein lysate, 1.5–2 mg was used per immunoprecipitation (IP) in IP buffer (10 mM Tris-HCl pH 7.6, 150 mM NaCl and 0.2% NP-40) supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (78445, Thermo Fisher Scientific). Of IP volume, 10% was designated for input assessments. Protein lysate was immunoprecipitated with 10 µg of antibody pre-bound to 30 µl of washed protein G Dynabeads (Invitrogen) per IP. IPs were conducted overnight at 4 °C, washed three times with IP buffer and once with the IP wash Buffer (10 mM Tris-HCl pH 7.6, 250 mM NaCl and 0.2% NP-40), both supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (78445, Thermo Fisher Scientific). Then IPs were eluted in Laemmli buffer by boiling, then isolated from beads and transferred to new tubes for western blot analysis.
Software and statistical analysis
All experiments were performed with at least three replicates, with the specific number of replicates stated in the figure legends. Unless otherwise stated, statistical analyses were performed using GraphPad prism (v10.3.0) using two-way ANOVA, and statistical significance was determined at a P < 0.05.
Related to Fig. 5a–c, note that in the boxplots, the middle line represents the median, the lower and upper hinges represent the 25th and 75th percentiles, respectively, the lower whisker extends from the lower hinge to the smallest value at most 1.5× the interquartile range of the hinge, and the upper whisker extends from the upper hinge to the largest value no further than 1.5× the interquartile range of the hinge. Data beyond the whiskers are outliers that are plotted individually.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.