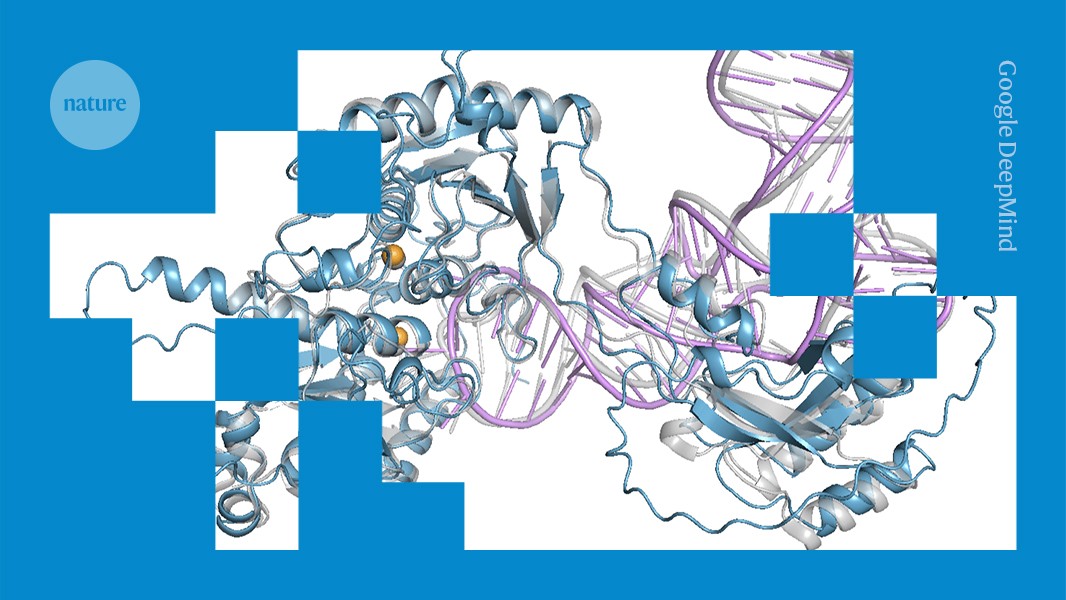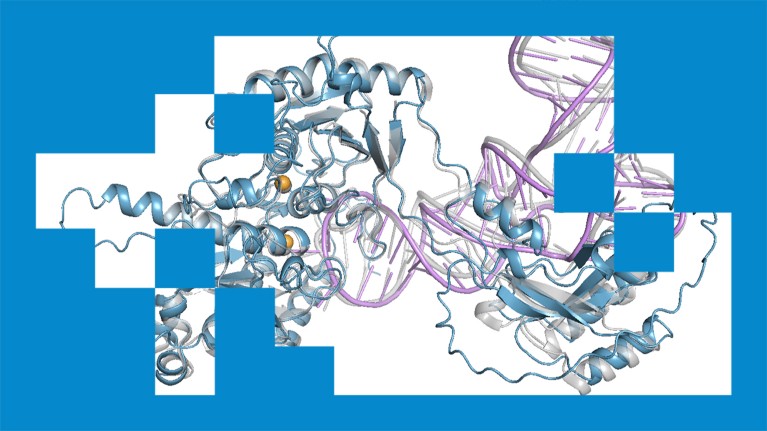
An AlphaFold3 model of a protein (blue) interacting with RNA (purple).Credit: Google DeepMind
Scientists today released a ‘sneak preview’ of a new open-source artificial intelligence (AI) model that predicts 3D structures of proteins, and say it is close to matching the performance of Google DeepMind’s revolutionary protein-folding AI AlphaFold3.
The system, called OpenFold3, was developed by the OpenFold Consortium1, a non-profit collaboration of academic and private research groups, headquartered in Davis, California. OpenFold3 uses proteins’ amino acid sequences to map their 3D structures and model how they interact with other molecules, such as drugs or DNA.
The tool still doesn’t have the same functionality as AlphaFold3, but “we wanted to get something out to the community as soon as possible”, says Woody Sherman, the consortium’s executive committee chair and chief innovation officer at the firm Psivant Therapeutics in Boston, Massachusetts. The team hopes to use researcher feedback after the preview release to improve the model.
Full release coming
The system was trained on more than 300,000 molecular structures and a synthetic database of more than 40 million structures. Developing it has cost US$ 17 million so far.
Who will make AlphaFold3 open source? Scientists race to crack AI model
Unlike AlphaFold3, which is available for restricted academic use, any researcher or pharmaceutical company can use OpenFold3. “It’s a big step forward in terms of the democratization of AI structural-biology tools,” says Sherman.
The preview release shares OpenFold3’s code with researchers and allows them to start using and testing it. Sherman says that the consortium team is still working on technical improvements to the system and is planning a full release in the coming months. “We’re close — and we think, with this last little bit of effort, we’re going to get fully to parity,” he says. “This is a way for folks to get a flavour of OpenFold3 and start integrating it into their workflows and start building tools and start providing feedback.”
“I’m excited to test [OpenFold3] and see how [it] compares to existing models,” says Stephanie Wankowicz, a computational structural biologist at Vanderbilt University in Nashville, Tennessee.
Open-source models
OpenFold3 is part of a wider push to develop open-source versions of AlphaFold3. These efforts began in May 2024, when London-based Google DeepMind launched AlphaFold3 without initially sharing its underlying code. Researchers criticized the private firm’s decision — and later, in November 2024, DeepMind made the AlphaFold3 code and model weights, or all the trainable parameters in an AI model, available to academics (they remain unavailable for commercial use).