Mice
All experiments conducted in this study were approved by the MIT Committee on Animal Care or the MGH Institutional Animal Care and Use Committee. Female B6 and female NSG mice 6–10 weeks of age were used. Animals were housed at ambient temperature and humidity (18–23 °C, 40–60% humidity) on a 12-h light–12-h dark cycle and co-housed with littermates with ad libitum access to food and water. All experimental groups were age matched, numbered and randomly assigned to treatment or control groups to minimize bias. Randomization was performed using a randomized numbering sequence at the time of injection or treatment. Investigators were not blinded during animal injections or treatments; however, researchers were blinded during data acquisition and quantification. No statistical methods were used to pre-determine sample size. Animal studies were performed once per condition unless otherwise specified in the figure legends. Data were collected from distinct animals, where n represents biologically independent samples. Mice were euthanized when total tumour burden reached 2 cm in diameter or earlier if humane end point criteria (for example, more than 20% weight loss, body condition score of less than 2 or signs of distress) were observed. These institutional limits were not exceeded in any experiment.
Cell lines and culture
Human cell lines used in this study were obtained from the American Type Culture Collection (HCC1806: CRL-2335, from a female patient; MDA-MB-231: HTB-26, from a female patient). Mouse EO771 breast cancer cells were obtained from CH3 BioSystems (94A001, from B6 mice). Cell lines obtained from ATCC and CH3 BioSystems are authenticated by the suppliers; no additional authentication was performed by the authors. Lines were regularly tested for mycoplasma contamination using the MycoAlert PLUS Mycoplasma Detection Kit (LT07-710, Lonza BioSciences). All cells were cultured in a Heracell humidified incubator (Thermo Fisher Scientific) at 37 °C and 5% CO2. Cell lines were routinely maintained in RPMI-1640 (10-040-CV, Corning Life Sciences) supplemented with 10% heat-inactivated fetal bovine serum (10437-028, Gibco), and for cell culture experiments, 10% dialysed fetal bovine serum (26400-044, Gibco) was supplemented.
RPMI-1640 medium lacking serine or arginine was made using a previous method61. In brief, enough of all amino acids and sodium phosphate dibasic (S5136, Sigma-Aldrich) of RPMI-1640 media except for serine or arginine were weighed out to make 25 l of media, then the resulting powders were homogenized using an electric blade coffee grinder (80365, Hamilton Beach) that had been washed with methanol then water. The resulting powders were resuspended in water along with sodium bicarbonate (S5761, Sigma-Aldrich) and RPMI-1640 medium without amino acids and sodium phosphate (R8999-04A, US Biological) to make RPMI-1640 medium lacking serine or arginine. Serine or arginine was reconstituted in water to generate stock solutions and added back as needed to reach 286 µM serine or 1.15 mM arginine, reflecting standard RPMI-1640 concentrations.
DMEM medium lacking serine or arginine was prepared using a base powder formulation lacking all amino acids and pyruvate (D9800-13, US Biological). To this base, glucose, sodium pyruvate and sodium bicarbonate were added to match standard DMEM composition. Amino acids were weighed out and homogenized as described above for RPMI, then dissolved into the reconstituted DMEM base. Serine or arginine was added back as needed to reach 400 µM serine or 398 µM arginine, reflecting standard DMEM concentrations.
Isolation of interstitial fluid, CSF and plasma
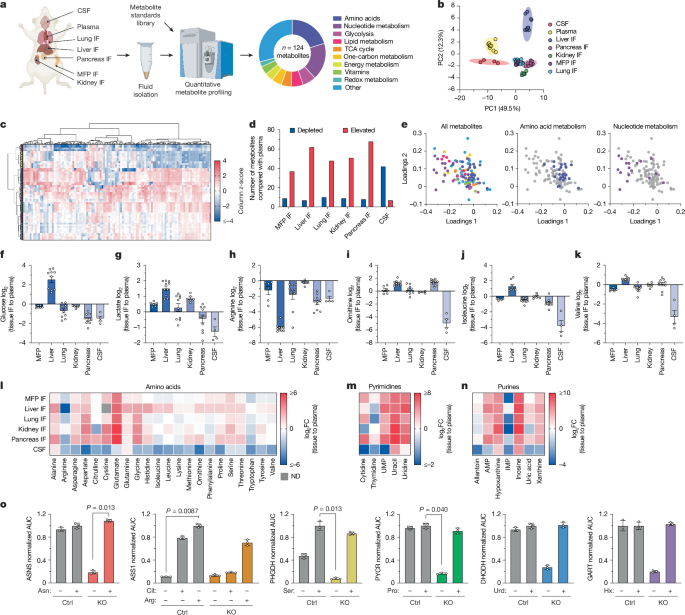
Tissue interstitial fluid was collected from mouse tissues using an adapted protocol based on previous published work52. Female NSG or female B6 mice on ad libitum diet 6–9 weeks of age were used for all tissue interstitial fluid isolations. Organs from five mice were combined per interstitial fluid sample, and each pooled sample was treated as an individual data point for the liquid chromatography–mass spectrometry (LC–MS) measurements and analysis. All mice were euthanized at the same time of day to account for any effects of time of day on metabolism, and were euthanized by cervical dislocation to minimize metabolic artefacts associated with CO2 exposure62. Tissues were kept on a chilled metal block on ice throughout the harvest and when ready to pool, were briefly rinsed in ice-cold saline (16005, Azer Scientific), excess liquid removed by blotting on filter paper (28298-020, VWR), then placed in a 50-ml conical vial lined with a 20-μm nylon mesh filter (148134, Spectrum Labs) and 0.25 μl of 0.5 M EDTA, pH 8.0 at the bottom. The tissues were centrifuged at 400g for 10 min at 4 °C. The flow through was collected and centrifuged again at 400g for 10 min at 4 °C before flash freezing and storage in −80 °C until further analysis. Before tissue harvest, plasma from each live mouse was collected by facial cheek bleed into EDTA-coated tubes (41.1395.105, Sarstedt), then centrifuged at 800g for 10 min at 4 °C. Supernatant containing plasma was flash frozen and stored at −80 °C until further analysis. An equal volume of each plasma sample was pooled from each mouse cohort before analysis by LC–MS.
CSF was isolated from the mouse brain as previously described63. In brief, female NSG or female B6 mice were anaesthetized with ketamine (90 mg kg−1) and xylazine (9 mg kg−1) via intramuscular injection, and CSF was collected by gently inserting a sharpened capillary (inner diameter of 0.75 mm and outer diameter of 1.0 mm) in the cisterna magna. Collected fluid was visually monitored for blood contamination. CSF was centrifuged at 800g for 10 min at 4 °C, and the supernatant was flash frozen and stored at −80 °C until further analysis. Following CSF collection, animals were euthanized via cervical dislocation and blood was collected via cardiac puncture and immediately placed in EDTA tubes (41.1395.105, Sarstedt), then centrifuged at 800g for 10 min at 4 °C. Plasma was flash frozen in liquid nitrogen and stored at −80 °C until further analysis.
To evaluate the stability of metabolite levels during sample handling, we performed a time-course experiment in female B6 mice. For plasma, blood was collected via cheek bleed directly into EDTA-coated tubes and either processed immediately as described above or held on ice for 5, 10 or 30 min before processing. For liver interstitial fluid, one liver was collected from each mouse and either processed immediately or placed on a chilled metal block on ice for 5, 10 or 30 min before interstitial fluid isolation using the standard centrifugation protocol described above. All samples were analysed by LC–MS to quantify time-dependent changes in metabolite levels.
MFP and brain tumour model generation
MDA-MB-231, HCC1806 and EO771 cell lines were transduced with a lentiviral vector expressing Gluc and GFP separated by an internal ribosomal entry site42, and the top 10% of GFP+ cells were isolated by FACS. To generate MFP tumours, mice were first anaesthetized with ketamine (90 mg kg−1) and xylazine (9 mg kg−1) via intraperitoneal injection, and 1 × 105 MDA-MB-231–Gluc or HCC1806–Gluc cells were injected into the MFP in 30 or 50 µl volume of PBS. To generate brain tumours, mice were first anaesthetized with ketamine (90 mg kg−1) and xylazine (9 mg kg−1) via intramuscular injection. Then, 1 × 105 MDA-MB-231–Gluc, 1 × 105 HCC1806–Gluc or 5 × 104 EO771–Gluc cells were diluted in 1 µl PBS and stereotactically injected into the left frontal lobe of the mouse brain. Tumour volumes were assessed 1–2 times per week by mixing 7 µl tail vein blood with 7 µl 0.5 mM EDTA (pH 8.0) and quantifying luminescence using Promega GloMax Plate Reader (Promega) and the substrate coelenterazine (303, NanoLight Technology).
Intracardiac injection and metastasis quantification
MDA-MB-231, HCC1806 and EO771 cell lines were transduced with a lentiviral vector expressing Fluc and GFP12, and infected cells were sorted by FACS using a fixed gate for GFP. For intracardiac injections, 1 × 105 cancer cells were suspended in 100 µl PBS and injected into the left ventricle of mice anaesthetized with inhaled isoflurane using ultrasound guidance64. In vivo metastasis progression was monitored via real-time bioluminescence imaging (BLI) using the IVIS Spectrum Imaging System (PerkinElmer). Mice were anaesthetized with inhaled isoflurane, injected intraperitoneally with D-luciferin (150 mg kg−1), then imaged every 3 min with the auto-exposure setting in supine position for a total time of 15 min. The time at which luminescence reached its maximum was used for total photon flux values (photons per second).
For end point ex vivo tissue BLI quantification, mice were intraperitoneally injected with D-luciferin (150 mg kg−1), anaesthetized with inhaled isoflurane and whole-body BLI was recorded 10 min post-luciferin injection. Following immediate euthanasia by cervical dislocation, tissues were rapidly dissected and imaged with the auto-exposure setting. BLI analysis was conducted using Living Image software (v4.7.2, PerkinElmer).
Generation of metastatic cell lines
Following intracardiac injection of GFP-labelled or Fluc-labelled cancer cells and end point BLI, GFP+ metastases in the brain and/or liver were identified and microdissected using a fluorescent dissecting microscope. Brain metastases were placed directly into 15-ml conical tubes containing 5 ml of digestion buffer (1 mg ml−1 collagenase I (LS004194, Worthington Biochemical), 3 mg ml−1 dispase II (04942078001, Roche) and 0.1 mg ml−1 DNase I (D4527, Sigma) in PBS). Liver tissue bearing GFP+ metastases were placed into digestion buffer and finely minced using sterile surgical tools. Tissues were then digested at 37 °C for 1 h with gentle rotation. After digestion, samples were triturated using a flamed glass Pasteur pipette to further dissociate tissue. EDTA was added to a final concentration of 10 mM, and samples were incubated at room temperature for 5 min. Cell suspensions were filtered through a 70-µm cell strainer into 50-ml conical tubes, centrifuged, washed twice with PBS and resuspended in RPMI medium supplemented with 10% heat-inactivated fetal bovine serum and penicillin–streptomycin. Cells were plated and expanded in culture, and GFP+ populations were subsequently isolated by flow cytometry for downstream analyses.
Petal plot generation
To account for baseline activity and inter-tissue variability in our luciferase assay data, we normalized the readings against control cell lines expressing non-targeting control single guide RNA. This normalization involved dividing the raw data for each gene by the average non-targeting control values corresponding to each tissue type. We used bootstrap methods for statistical reliability, resampling the normalized data to estimate mean value distributions and calculate 95% confidence intervals, thereby capturing the inherent variability in the biological data. Values indicating fold changes above 1, suggesting normal or enhanced cell growth, were capped at 1 to focus on growth dependencies. Data visualization, inspired by Jin et al.12, was achieved through a petal plot (a radial variant of a bar plot), succinctly depicting luciferase activities along with 95% confidence intervals. Replicates within the dataset were averaged to provide a single representative measure per tissue type. The analysis was conducted using R studio (v4.3.1), using the dplyr (v1.1.2) and ggplot2 (v3.4.3) packages for data processing and visualization, the boot package (v2019.6.0) for bootstrap confidence interval calculations, and the reshape2 package (v1.4.4) for data reshaping.
Quantification of metabolite levels in biological fluids
Metabolite quantification in mouse fluid samples was performed as previously described52. In brief, 5 μl of sample or external chemical standard pool (ranging from approximately 5 mM to about 1 μM) was mixed with 45 μl of acetonitrile:methanol:formic acid (75:25:0.1) extraction mix including isotopically labelled internal standards. All solvents used in the extraction mix were high-performance LC grade (HPLC). Samples were vortexed for 15 min at 4 °C, and insoluble material was sedimented by centrifugation at 16,000g for 10 min at 4 °C. Of the soluble polar metabolite extract, 20 µl was taken for LC–MS analysis. Following analysis by LC–MS, metabolite identification was performed with XCalibur 2.2 software (Thermo Fisher Scientific) using a 5 ppm mass accuracy and a 0.5-min retention time window. For metabolite identification, external standard pools were used for assignment of metabolites to peaks at given m/z and retention time (see Supplementary Table 1 for the m/z and retention time for each metabolite analysed). Absolute metabolite concentrations were determined as previously published52.
13C-glucose tracing in brain and MFP tumours
Infusion of [U-13C]-glucose (CLM-1396-1, Cambridge Isotope Laboratories) into tumour-bearing mice was performed as previously described13,65,66. MDA-MB-231-Gluc cells (1 × 105) were intracranially injected into the brain or injected into the MFP of mice and tumours were permitted to grow for 14 or 17 days, respectively. Catheters were surgically implanted into the jugular vein of tumour-bearing animals 3 days before the infusion. Mice were fasted for 4 h before the start of the infusion, then conscious, free-moving animals were infused with [U-13C]-glucose at a constant rate of 0.4 mg min−1 for 10 h. Mice remained fasted throughout the infusion period. Following the 10-h infusion, animals were terminally anaesthetized with an infusion of Fatal Plus. Blood was collected immediately by cardiac puncture, and tumours and non-cancerous tissue were collected within 5 min of death; for brain tumours, GFP+ tumour tissue was isolated from non-GFP+ tissue under a fluorescent dissecting microscope. Tissues were flash frozen using the BioSqueezer (1210, BioSpec Products) and stored at −80 °C for subsequent metabolite extraction and analysis. Blood was placed into EDTA tubes (41.1395.105, Sarstedt) and centrifuged at 800g at 4 °C to separate plasma, which was flash frozen and stored at −80 °C. All isotope labelling experiments in mice were performed at the same time of day.
Snap frozen tissues were ground into powder using a mortar and pestle, then weighed into glass vials (C4010-1 and C4010-60BLK, Thermo Fisher Scientific). Metabolites were extracted in 1.5 ml of dichloromethane:methanol:0.88% KCl (w/v) (8:4:3 v/v/v), vortexed for 15 min at 4 °C and centrifuged at maximum speed for 10 min at 4 °C. Polar metabolites (aqueous fraction) were transferred to Eppendorf tubes, dried under nitrogen gas and stored at −80 °C until further analysis. Before analysis by LC–MS, samples were resuspended in water, vortexed for 10 min, centrifuged at maximum speed for 10 min and supernatant transferred to vials. For plasma, 5 µl of fluid was extracted in 45 µl of 80% methanol containing 500 nM 13C and 15N amino acid standards (MSK-A2-1.2, Cambridge Isotope Laboratory). Samples were vortexed for 15 min, centrifuged at maximum speed for 10 min, and supernatant transferred to vials and analysed by LC–MS. Following analysis by LC–MS, metabolite peak areas were called using XCalibur (v.2.2; Thermo Fisher Scientific) or Compound Discoverer (v.3.3; Thermo Fisher Scientific) software. Ion counts for each metabolite were normalized to the weight of the tissue sample; for plasma samples, total ion counts were reported. Isotope corrections were applied using IsoCorrector (v.3.18)67.
Dietary serine or glycine restriction and metastasis assay
To assess the effect of systemic nutrient availability on metastasis, female NSG mice (6–8 weeks of age) were fed either a control diet (TD.110839, Envigo) or a high-serine diet (TD.160575, Envigo). Mice were maintained on the assigned diets for 2 weeks before intracardiac injection of GFP-lablled or Fluc-labelled HCC1806 cells. Mice were maintained on each diet until end point. For metabolomics analysis of tissues of mice on the diets, separate mice were placed on the two diets for 2 weeks at which point plasma and tissues were collected and analysed by LC–MS. Mice were randomly assigned to diets, and all tissue and plasma samples were collected at the same time of day to control for circadian effects. Before tissue harvest, plasma from each live mouse was collected by facial cheek bleed into EDTA-coated tubes (41.1395.105, Sarstedt), then centrifuged at 800g for 10 min at 4 °C. Supernatant containing plasma was flash frozen and stored at −80 °C until further analysis. Tissues were rinsed in cold saline, blotted dry, then snap frozen and stored at −80 °C. Frozen tissues were ground into powder using a mortar and pestle and metabolites were extracted in 50 µl extraction mix (80% methanol + containing 500 nM 13C and 15N amino acid standards (MSK-A2-1.2, Cambridge Isotope Laboratory)) per mg of tissue. Samples were vortexed for 15 min, centrifuged at maximum speed for 10 min, dried down under nitrogen gas and then stored at −80 °C. For analysis, samples were resuspended in HPLC-grade water and analysed by LC–MS.
For plasma, 5 µl of fluid was extracted in 45 µl of 80% methanol containing 500 nM 13C and 15N amino acid standards (MSK-A2-1.2, Cambridge Isotope Laboratory). Samples were analysed by quantitative metabolomics as described below. Samples were vortexed for 15 min, centrifuged at maximum speed for 10 min, and supernatant was transferred to vials and analysed by LC–MS. Following analysis by LC–MS, metabolite peak areas were called using XCalibur (v.2.2; Thermo Fisher Scientific).
In vitro [U-¹³C]-glucose tracing
MDA-MB-231 control or DHODH-knockout cells were seeded in six-well plates in RPMI supplemented with 100 µM uridine. After attachment, cells were washed and incubated for 24 h in RPMI (R8999, US Biological) containing 10 mM [U-13C6]-glucose (CLM-1396-1, Cambridge Isotope Laboratories) and 100 µM uridine. Following incubation, cells were washed, extracted in 80% methanol containing 500 nM 13C and 15N amino acid standards (MSK-A2-1.2, Cambridge Isotope Laboratory), vortexed, clarified by centrifugation, dried and stored at −80 °C until analysis by LC–MS. Replicate plates were counted in parallel, and metabolite peak areas were normalized to internal standards and to cell number.
LC–MS analysis
Metabolite profiling was conducted on a QExactive benchtop orbitrap mass spectrometer equipped with an Ion Max source and a Heated Electrospray Ionization (HESI-II) probe, which was coupled to a Dionex UltiMate 3000 HPLC system (Thermo Fisher Scientific). External mass calibration was performed using the standard calibration mixture every 7 days. An additional custom mass calibration was performed weekly alongside standard mass calibrations to calibrate the lower end of the spectrum (m/z 70–1,050 positive mode and m/z 60–900 negative mode) using the standard calibration mixtures spiked with glycine (positive mode) and aspartate (negative mode). Of each sample, 2 µl was injected onto a SeQuant ZIC-pHILIC 150 × 2.1 mm analytical column equipped with a 2.1 × 20 mm guard column (both particle size of 5 mm; EMD Millipore). Buffer A was 20 mM ammonium carbonate and 0.1% ammonium hydroxide; and buffer B was acetonitrile. The column oven and autosampler tray were held at 25 °C and 4 °C, respectively. The chromatographic gradient was run at a flow rate of 0.150 ml min−1 as follows: for 0–20 min, linear gradient from 80% to 20% B; for 20–20.5 min, linear gradient from 20% to 80% B; and from 20.5–28 min, hold at 80% B. The mass spectrometer was operated in full-scan, polarity-switching mode, with the spray voltage set to 3.0 kV, the heated capillary held at 275 °C and the HESI probe held at 350 °C. The sheath gas flow was set to 40 units, the auxiliary gas flow was set to 15 units and the sweep gas flow was set to 1 unit. MS data acquisition was performed in a range of m/z = 70–1,000, with the resolution set at 70,000, the automatic gain control target at 1 × 106 and the maximum injection time at 20 ms.
Cell proliferation
For assessment of proliferation by continuous live-cell imaging, cells were trypsinized, pelleted and washed with PBS, counted, resuspended in the appropriate medium and then plated directly into clear 96-well plates. Cells were then treated with or without the relevant rescue metabolite, and plates were placed into an IncuCyte Live Cell Analysis Imaging System S3 (Sartorius) in a humidified incubator at 37 °C and 5% CO2. Images were acquired every 3 h using the 10x objective. Cell confluence was determined from a mask generated by the standard settings in the IncuCyte Zoom Analysis S3 v2018B software. Cells were plated in either RPMI or DMEM medium lacking the metabolite corresponding to their engineered auxotrophy: asparagine (ASNS knockout), arginine (ASS1 knockout), proline (PYCR knockout) or serine (PHGDH knockout). DHODH-knockout and GART-knockout experiments were performed in complete RPMI medium. The metabolite concentrations used for rescues unless otherwise stated were: 1.15 mM arginine, 379 µM asparagine, 1 mM citrulline, 100 µM hypoxanthine, 174 µM proline, 286 µM serine and 100 µM uridine.
Western blot
Cells were washed once in PBS and scraped into RIPA lysis buffer containing protease and phosphatase inhibitors (5871, Cell Signaling Technology), rocked for 15 min at 4 °C, and insoluble material was sedimented by centrifugation at 21,000g for 10 min at 4 °C. Protein concentration was determined via Bradford Assay, and samples were mixed with LDS sample buffer (NP0008, Thermo Fisher Scientific) and 2.5% 2-mercaptoethanol and then incubated at 95 °C for 5 min. Proteins were resolved by SDS–PAGE and then transferred onto nitrocellulose membranes using a wet tank transfer system (Bio-Rad). Membranes were blocked in 5% milk in TBST and probed overnight at 4 °C with the appropriate antibody diluted in 5% BSA in TBST. For detection, membranes were incubated with horseradish peroxidase-linked anti-rabbit (1:5,000; 7074, Cell Signaling Technology) or anti-mouse (1:5,000; 7076, Cell Signaling Technology) IgG secondary antibody diluted in 5% milk in TBST, and chemiluminescent signal was detected using a digital imager (LAS 4000, GE Healthcare). The primary antibodies used were: ASNS (1:1,000; 14681-1-AP, Proteintech), ASS1 (1:1,000; clone D4O4B, 70720, Cell Signaling Technology), PHGDH (1:1,000; HPA021241, Sigma-Aldrich), PYCR1 (1:1,000; 13108-1-AP, Proteintech), PYCR2 (1:1,000; 17146-1-AP, Proteintech), PYCRL (1:1,000; clone OTI1B12, NBP2-03337, Novus Biologicals), PYCRL (1:1,000; A17763, ABclonal), DHODH (1:100; clone E-8, sc-166348, Santa Cruz Biotechnology), GART (1:1,000; 13659-1-AP, Proteintech), β-actin (1:5,000; clone D6A8, 8457, Cell Signaling Technology) and vinculin (1:1,000; clone E1E9V, 13901, Cell Signaling Technology). In cases in which re-probing of the same membrane was required, the membrane was incubated with hydrogen peroxide solution 30% (w/w in water; H1009, Sigma-Aldrich) for 1 h at 37 °C to inactivate horseradish peroxidase. Western blots were quantified by densitometry using Fiji (v2.0.0)68; data are normalized to the loading control from the same gel. Uncropped, unprocessed scans for all key blots are provided in Supplementary Fig. 1.
Lentivirus generation
LentiX cells were transfected with lentiviral packaging plasmids pMD2.G (12259, Addgene) pMDLg/pRRE (12251, Addgene) and pRSV-Rev (12253, Addgene) using TransIT-293 Transfection Reagent (MIR 2700, Mirus). Forty-eight hours post-transfection, medium was collected and filtered through a 0.45-µm low-protein-binding membrane. Lentivirus-containing supernatants were used immediately or stored at −80 °C. Cell lines were transduced with virus and 10 µg ml−1 polybrene for 24 h.
CRISPR–Cas9-mediated gene knockout
Cas9-expressing cell lines were generated by infecting parental cell lines with viral particles containing lentiCas9/blast (52962, Addgene) followed by selection with 10 µg ml−1 blasticidin. Two guide RNAs (gRNAs) per gene of interest were designed by CRISPick (https://portals.broadinstitute.org/gppx/crispick/public) and cloned into lentiGuide-Puro (52963, Addgene). Primer sequences used in this study are listed in Supplementary Table 3. Cas9+ cell lines were transduced with lentiGuide-Puro-gRNA viral particles, then selected with 1 µg ml−1 puromycin. Single cells were plated into individual wells of 96-well plates in RPMI medium; following expansion, western blotting confirmed complete knockout. To generate and maintain DHODH-knockout or GART-knockout cells, 100 µM uridine or 100 µM hypoxanthine were supplemented in RPMI medium, respectively. For the generation of PYCR1, PYCR2, PYCR3 (PYCRL) triple knockout MDA-MB-231 or HCC1806 cells, two gRNAs targeting both PYCR1 (exons 4 and 5) and PYCR2 (exons 2 and 3), and two gRNAs targeting PYCR3 (exon 3), were used.
To knock out Pycr1, Pycr2 and Pycr3 (PycrI) in EO771, we used ribonucleoprotein (RNP) complexes with Cas9–GFP and Cas9–RFP. The Cas9 protein and gRNAs specific to each target gene were ordered from Integrated DNA Technologies (IDT) as RNA oligonucleotides and used to assemble RNP complexes. EO771 cells were nucleofected with the RNP complexes using a 4D-Nucleofector (Lonza Bioscience) and the SF Cell Line 4D-Nucleofector X Kit L (V4XC2024, Lonza Walkersville) with Pulse Code CM-150. Post-nucleofection, cells were cultured and monitored for GFP and RFP expression. Single-cell sorting was performed 24 h after nucleofection to isolate successfully nucleofected cells.
DepMap and metastatic potential data
Gene expression, CRISPR-gene dependency and metastatic potential values used for correlation analyses were sourced from the Broad Institute DepMap portal website, 24Q2 release (https://depmap.org/portal). Metastatic potential values were derived from Jin et al.12, which used a standardized in vivo barcoding approach to assess tissue-specific metastatic behaviour across a large panel of human cancer cell lines.
Statistics and reproducibility
All in vitro experiments were independently repeated at least twice with similar results. Mouse experiments were performed once for each condition unless otherwise specified. All n values represent biologically independent samples. Sample sizes, reproducibility and statistical tests used for each figure are denoted in the figure legends or Source Data. All graphs were generated using GraphPad Prism 10.3.1 (GraphPad Software). Relative fold changes were determined by normalizing every data point to the mean value of the control group.
After determining the concentration of each metabolite in each plasma or tissue interstitial fluid sample, all multivariate statistical analysis on the data was performed using Metaboanalyst (v6.0)69. Metabolite concentrations were log transformed, and metabolites that contained greater than 50% missing values were removed before analysis. For the remainder of the metabolites, missing values were replaced using one-fifth of the lowest positive value. Hierarchical clustering was performed with Euclidean distance measurement and clustering by the Ward algorithm. Univariate analysis was performed comparing metabolite levels between groups in which metabolite differences of interest were defined by a fold change greater than 2 and significance as a raw P value of less than 0.05 assuming unequal group variance. Using the fold change and adjusted P value cut-offs, the number of differentially expressed metabolites was determined.
Illustrations
Experimental schema and illustrative models were generated using BioRender.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.