Purification of wild-type SWR1
Recombinant SWR1 was produced as previously described12,16 with minor modifications. Baculoviruses encoding SWR1 genes were initially amplified in Sf9 cells, before using the amplified baculoviruses to infect BTI-TN-5B1-4 (High Five) cells for expression, which were harvested after 72âh. Cells were lysed by sonication in 50âmM HEPES (pH 8.0), 0.5âM NaCl, 1âmM TCEP, 10% glycerol, 1âmM benzamidine-HCL supplemented with 1 protease inhibitor tablet and 10âµl of benzonase per litre of cell culture. Lysate was clarified by centrifugation at 30,000g for 60âmin at 4â°C. The supernatant was filtered before being injected onto a StrepTrap HP (Cytiva) column. The column was washed with buffer A (25âmM HEPES (pH 7.5), 0.3âM NaCl, 1âmM TCEP and 10% glycerol) before being eluted with buffer A supplemented with 5âmM desthiobiotin. The eluted protein was combined and diluted 1:1 with buffer B (25âmM HEPES (pH 7.5), 0.1âM NaCl, 1âmM TCEP and 10% glycerol) to dilute the salt before being loaded onto a HiTrap Q HP (Cytiva) column. The protein was eluted with a linear gradient from buffer B to buffer C (25âmM HEPES (pH 7.5), 2âM NaCl, 1âmM TCEP and 10% glycerol). The relevant fractions were pooled and diluted again 1:1 with buffer B to reduce the salt before being injected onto a Heparin HP (Cytiva) column. Protein was eluted with a linear gradient from buffer B to buffer C. Finally, the protein was concentrated, snap frozen in liquid nitrogen and stored at â80â°C.
Purification of fluorescently labelled SWR1
To site specifically label the SWR1 complex, we made use of the ybbR-labelling approach34,35. The 11-amino acid ybbR tag was fused to the N terminus of the Arp6 subunit of SWR1. The ybbRâArp6 mutant was used in place of the wild-type Arp6 gene when assembling the SWR1 genes using the MultiBac system16. The SWR1(ybbRâArp6) complex was expressed and purified in an analogous way to wild-type SWR1 with the ybbR-labelling reaction taking place after elution from the HiTrap Q HP column. The labelling reaction was carried out overnight at 4â°C. Typically, SWR1(ybbRâArp6; approximately 1âµM) was labelled with CoA-Atto647N (approximately 10âµM) using recombinant Sfp transferase (approximately 0.2âµM) in buffer B supplemented with 10âmM MgCl2. The labelled SWR1 complex was separated from free dye and Sfp transferase using a Heparin HP (Cytiva) column, eluting with a linear gradient from buffer B to buffer C. Finally, SWR1(Atto647NâArp6) (referred to as SWR1(647N) in the text) was concentrated, snap frozen in liquid nitrogen and stored at â80â°C.
Purification of S. cerevisiae histones
All nucleosomes or hexasomes used in this study were composed of S. cerevisiae histones assembled on DNA containing the 601 Widom sequence.
S. cerevisiae octamers with and without Alexa Fluor 555 on H2A K119C were prepared as previously described16.
S. cerevisiae H2AâH2B, Htz1âH2B (with and without Alexa Fluor 555 on Htz1 K125C) or Htz1âH2B(3ÃFlag) histone dimers were expressed in E. coli and purified as soluble dimers. Cells were lysed by sonication in buffer D (20âmM Tris (pH 7.5), 0.5âM NaCl, 0.1âmM EDTA and 1âmM TCEP) plus protease inhibitor tablets (Roche; 2 tablets per 100âml). Dimers were purified by loading the cleared lysate onto tandem HiTrap Q FF and HiTrap Heparin HP columns in buffer E (20âmM Tris (pH 7.5), 0.5âM NaCl, 1âmM EDTA and 1âmM TCEP). The HiTrap Q FF column was removed before elution from the HiTrap Heparin HP column via a gradient to buffer F (20âmM Tris (pH 7.5), 2âM NaCl, 1âmM EDTA and 1âmM TCEP), followed by gel filtration on a Superdex S200 in buffer F.
S. cerevisiae histone H3(Q120M, K121P and K125Q) and histone H4 were co-expressed in E. coli and purified as soluble tetramers. Cells were lysed by sonication in buffer D plus protease inhibitor tablets (Roche; 2 tablets per 100âml). Tetramers were purified using a HiTrap Heparin HP column in buffer E and eluted via a gradient to buffer F, followed by gel filtration on a Superdex S200 in buffer F.
Preparation of nucleosomes
Biotinylated DNA containing the Widom 601 sequence was generated as previously described12. Salt gradient dialysis of the S. cerevisiae octamers with DNA was carried out to form a âcoreâ nucleosome. A biotinylated DNA overhang was ligated to the core nucleosome as previously described12. This resulted in nucleosomes with one long overhang of 113âbp and a short overhang of 2âbp, which we refer to as 113N2 (âNâ representing the Widom 601 nucleosome positioning sequence). The biotin was present on the long 113-bp linker. For nucleosomes where the DNA was labelled, the fluorophore was attached at the end of the 2-bp short overhang.
Preparation of hexasomes
To facilitate the formation of yeast hexasomes, three amino acid substitutions were introduced into the S. cerevisiae H3 histone (Q120M, K121P and K125Q)36. These substitutions (MPQ) are the corresponding amino acids found in human and Xenopus laevis H3.
To form hexasomes, S. cerevisiae H2AâH2B dimers were mixed with S. cerevisiae H3(MPQ)âH4 tetramers. The amount of H2AâH2B dimers used was limited to 0.6Ã the amount of tetramers to ensure only partial H2AâH2B occupancy. Hexasomes were assembled onto the same DNA that was used for nucleosomes by salt gradient dialysis to generate âcoreâ hexasomes. Core hexasomes were separated from tetrasomes, nucleosomes and free DNA using a MonoQ column, loaded in buffer G (20âmM Tris (pH 7.5), 1âmM EDTA, 1âmM TCEP and 200âmM NaCl) eluting with a gradient into buffer H (as buffer G with 2âM NaCl). The fractions were immediately diluted into 4Ã volume of 20âmM Tris (pH 7.5) to reduce the salt concentration. A biotinylated DNA overhang was ligated to the core hexasome in the same way as was used for nucleosomes. This resulted in a hexasome with one long overhang of 113âbp and a short overhang of 2âbp, which we refer to as 113H2 (âHâ representing a hexasome assembled on the Widom 601 sequence). The biotin was present on the long 113-bp linker. For hexasomes where the DNA was labelled, the fluorophore was attached at the end of the 2-bp short overhang.
As is the case for hexasomes prepared with X. laevis histones37, yeast hexasomes prepared in this way exploit the inherent asymmetry of the Widom 601 sequence. Because of this asymmetry, the H2AâH2B dimer present in a hexasome is preferentially located on the âTA-richâ side of the Widom 601 sequence, leaving the vacant site on the âTA-poorâ side. We orientated our Widom 601 sequence with the TA-rich side closest to the 2-bp short overhang. This resulted in the vacant H2AâH2B site being located next to the 113-bp linker.
Preparation of heterotypic nucleosomes
Core hexasomes, prepared as described above, were mixed with S. cerevisiae Htz1âH2B dimers to form heterotypic nucleosomes. Htz1âH2B dimers were added at an amount equal to 0.3Ã the amount of hexasome present. Core heterotypic nucleosomes were then purified in the same way as canonical nucleosomes. A biotinylated DNA overhang was ligated to the core heterotypic nucleosomes as described above. Resulting heterotypic nucleosomes contain the Htz1âH2B dimer next to the long 113-bp overhang and the conical H2AâH2B dimer next to the short 2-bp overhang.
Bulk histone exchange assay
SWR1 (100ânM; wild type or SWR1(647N)), 200ânM nucleosomes and 400ânM Htz1âH2B(3ÃFlag) were mixed in exchange buffer (25âmM Tris-HCl (pH 7.8), 100âmM KCl, 0.2âmM EDTA and 2âmM MgCl2), with or without 1âmM ATP. The exchange reaction was carried out at 30â°C. At the indicated time points, 8âµl of the reaction was removed and quenched by the addition of 4âµl of a stopping solution (0.5âmgâmlâ1 salmon sperm DNA, 30âmM EDTA and 3à ficoll loading buffer) and placed on ice. The âno ATPâ control was taken at the longest indicated time point. After all time points had been taken, the reaction products were separated by 6% native PAGE, run at 110âV in 0.5à TBE at 4â°C and visualized using fluorescence of the nucleosome.
Two-colour smFRET microscope
smFRET measurements looking at the flipping of nucleosomes by SWR1 were performed on an Olympus IX-71 microscope equipped with a homebuilt prism-TIRF module. Excitation was provided by a 532-nm laser (Stradus, Vortran) or a 637-nm laser (Stradus, Vortran). Fluorescence was collected through a 1.2 NA, 60à water objective (Olympus) and filtered through a dual bandpass filter (FF01-577/690-25, Semrock). The fluorescence was spectrally separated using a OptoSplit II (Cairn Research) to separate donor and acceptor emission. The donor and acceptor emissions were further filtered through ET585/65M and ET700/75M (Chroma) bandpass filters, respectively. The donor and acceptor images were then projected side-by-side onto an electron-multiplying charge-coupled device (EMCCD) (Andor iXon Ultra 897). Data were collected as raw movies using a custom LabVIEW script.
Single-molecule fluorescence spots from the raw movies were localized using custom IDL scripts and converted into raw fluorescence trajectories. Raw fluorescence trajectories were corrected for bleed through of the donor fluorescence into the acceptor channel. Apparent FRET efficiencies were calculated as the ratio of acceptor intensity divided by the sum of the donor and acceptor intensities.
Two mechanical shutters (LS-3, Uniblitz, Vincent Associates) were placed in the excitation path for alternating laser excitation (Extended Data Fig. 4eâg). Frame acquisition and shutter synchronization were obtained using a homebuilt negative-edge-triggered JK flipâflop circuit (SN74LS112AN, Texas Instruments) using the âFireâ output of the EMCCD as the input clock. IDL scripts were modified accordingly to locate single molecules and extract fluorescence trajectories.
Three-colour smFRET microscope
smFRET measurements looking at histone exchange coupled with SWR1 binding were performed on an Olympus IX-71 microscope equipped with a homebuilt prism-TIRF module. Alternating laser excitation was provided by a 488-nm laser (OBIS, Coherent) or a 637-nm laser (OBIS, Coherent). Alternation of the lasers and synchronization of the lasers with the camera were controlled by a custom LabVIEW script and a DAQ (USB-6341, National Instruments). Fluorescence was collected through a 1.2 NA, 60Ã water objective (Olympus) and filtered through ET500lp (Chroma) and NF03-642E-25 (Semrock) filters. The fluorescence was spectrally separated using a MultiSplit (Cairn Research) housing the following dichroic filters: T500lpxr UF2, T635lpxr UF2 and T725lpxr UF2 (Chroma). The separated fluorescent emission was projected onto quadrants of a sCMOS (ORCA Fusion, Hammamatsu) camera. Data were collected as raw movies using HCImage Live (Hammamatsu).
Single-molecule fluorescence spots from the raw movies were localized using custom IDL scripts and converted into raw fluorescence trajectories. Raw fluorescence trajectories were corrected for bleed through of the donor fluorescence into the acceptor channel. Apparent FRET efficiencies were calculated as the ratio of acceptor intensity divided by the sum of the donor and acceptor intensities.
Microscope slide passivation and flow chamber assembly
Quartz slides (UQC optics) and glass coverslips were aminosilinized with N-(2-aminoethyl)-3-aminopropyltrimethoxysilane, then passivated using methoxy-PEG-SVA (relative molecular massâ=â5,000; Laysan Bio, Inc.) containing 5% biotin-PEG-SVA (relative molecular massâ=â5,000, Laysan Bio, Inc.) in 100âmM sodium bicarbonate as previously described38 with minor modifications. Following passivation, slides and coverslips were stored under nitrogen in the dark at â20â°C. Before use, slides and coverslips were warmed to room temperature and assembled into flow chambers using 0.12-mm thick double-sided adhesive sheets (Grace Bio-Labs SecureSeal). Flow chambers were sealed with epoxy glue.
Nucleosome or hexasome immobilization
Nucleosomes or hexasomes were surface immobilized as previously described12. In brief, neutravidin (0.1âmgâmlâ1) in T50 buffer (50âmM Tris-HCl (pH 7.5) and 50âmM NaCl) was injected into the assembled flow chamber and incubated for 5âmin to allow binding to the biotinylated PEG surface. Excess neutravidin was washed out with reaction buffer (25âmM Tris-HCl (pH 7.8), 100âmM KCl, 4% glycerol, 1âmM EDTA, 2âmM MgCl2 and 0.2âmgâmlâ1 BSA). Biotinylated nucleosomes or hexasomes were diluted to 10âpM in reaction buffer before injecting into the flow chamber and allowed to bind to the neutravidin for 5âmin. Excess nucleosomes or hexasomes were flushed out using imaging buffer (reaction buffer with Trolox, 2.5âmM protocatechuic acid and 0.25âµM protocatechuate-3,4-dioxygenase) and imaged immediately.
smFRET between nucleosome or hexasome and SWR1 data collection
Nucleosomes or hexasomes labelled with a Cy3 donor on the short end of the DNA overhang (113N2.Cy3 or 113H2.Cy3) were immobilized in a flow chamber and imaged. SWR1(647N), 10ânM in imaging buffer (25âmM Tris-HCl (pH 7.8), 100âmM KCl, 4% glycerol, 1âmM EDTA, 2âmM MgCl2, 0.2âmgâmlâ1 BSA, Trolox, 2.5âmM protocatechuic acid and 0.25âµM protocatechuate-3,4-dioxygenase) was injected. Imaging was performed by first directly exciting the acceptor with a 637-nm laser for approximately 15âs to localize SWR1(647N), before switching to 532-nm excitation to observe FRET between the nucleosome or hexasome and SWR1. All single-molecule measurements were carried out at room temperature, data were acquired with a 100-ms frame time.
smFRET between nucleosome or hexasome and SWR1 data analysis
Manual inspection of the donor intensity, acceptor intensity and apparent FRET from each molecule was carried out using custom MATLAB scripts. For a molecule to be included in downstream analysis, it needed to have a constant signal from the acceptor under direct acceptor excitation to indicate that SWR1(647N) was bound and display a single step photobleaching event of either the donor or acceptor under donor excitation. All molecules that satisfied these criteria were truncated to just the FRETing region preceding the photobleaching event.
Truncated FRET traces were analysed with a hidden Markov model using vbFRET, using default parameters39. The idealized FRET from vbFRET was used to generate FRET histograms, plotted using Igor Pro 8 (Wavemetrics). Dwell times from the idealized FRET trajectories were extracted using custom MATLAB scripts. Only dwell times in which the idealized FRET transitioned between proximal and distal states (or the reverse) were included. Dwell time plots were generated in MATLAB and plotted in Igor Pro 8. The lifetime of the proximal-bound and distal-bound states was determined by fitting the dwell time plots to a double exponential function in Igor Pro 8. The slow and fast exponential phases probably correspond to a fully or partially engaged SWR1 complex, respectively. The average lifetimes (Ïave) for proximal-bound and distal-bound states were calculated using the pre-exponential factors (A) and lifetimes (Ï) determined from the double exponential fit as follows:
$${\tau }_{{\rm{ave}}}=({A}_{1}{{\tau }_{1}}^{2}+{A}_{2}{{\tau }_{2}}^{2})/({A}_{1}{\tau }_{1}+{A}_{2}{\tau }_{2})$$
In all cases, we observed both static and dynamic trajectories when probing the FRET between nucleosomes or hexasomes and SWR1. Only dynamic trajectories were used for determining the kinetics. For both the canonical and the heterotypic nucleosomes, static trajectories represent a minority of the observed molecules. Short static traces may be due to dye photobleaching or SWR1 diffusion before a flipping event can take place. However, longer static traces are also observed. This heterogeneity is summarized in Extended Data Fig. 5. Long static trajectories suggest that a proportion of SWR1 molecules are stably engaged on one side of the nucleosome and not dynamically checking the histone identity of each nucleosome face. The nature of this stable SWR1 binding, compared with binding that allows nucleosome flipping, is unknown, as is the method by which SWR1 could transition from a static (stable binding) to a flipping (checking histone identity) state.
smFRET real-time imaging of histone exchange and SWR1-binding data collection
A quartz flow cell was prepared as described above. Neutravidin (0.01âmgâmlâ1) in T50 buffer (50âmM Tris-HCl (pH 7.5) and 50âmM NaCl) was injected into the flow chamber and incubated for 5âmin to allow binding to the biotinylated PEG surface. Excess neutravidin was washed out and the flow cell further passivated by incubation with Pluronic F127 (0.5% w/v) in T50 buffer. Excess Pluronic F127 was washed out with reaction buffer (25âmM Tris-HCl (pH 7.8), 100âmM KCl, 4% glycerol, 0.2âmM EDTA, 2âmM MgCl2 and 0.2âmgâmlâ1 BSA).
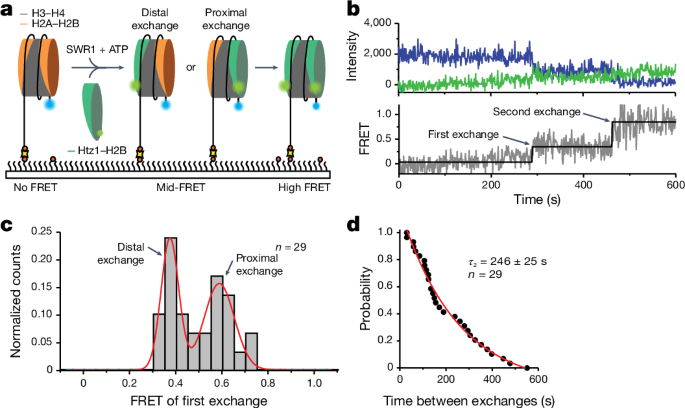
To follow the insertion of variant histones in real time at the single-molecule level, a âgain of FRETâ assay was used. Nucleosomes labelled with Alexa Fluor 488 (FRET donor) on the short 2-bp overhang (113N2.AF488) were immobilized in a flow chamber and imaged. To start the reaction, 1ânM SWR1, 4ânM Chz1âHtz1(AF555)âH2B and 1âmM ATP in imaging buffer (25âmM Tris-HCl (pH 7.8), 100âmM KCl, 4% glycerol, 0.2âmM EDTA, 2âmM MgCl2, 0.2âmgâmlâ1 BSA, Trolox, 2.5âmM protocatechuic acid and 0.25âµM protocatechuate-3,4-dioxygenase) was injected into the chamber using a syringe pump. Exchange can be monitored by stepwise FRET increases as the AF555-labelled (FRET acceptor) Htz1âH2B dimer is exchanged into the immobilized AF488-labelled nucleosome. To reduce nonspecific binding of the Htz1(AF555)âH2B dimer, the dimer was first complexed with its natural chaperone, Chz1 (ref. 40).
For experiments that simultaneously followed exchange and SWR1 binding, the experiment was conducted as described but with SWR1(647N) using the three-colour smFRET microscope described above. The two excitation lasers (488ânm and 637ânm) were alternated at a frequency of 1âHz. All experiments were carried out at room temperature (22â°C).
smFRET real-time imaging of histone exchange and SWR1-binding data analysis
Visualization of single-molecule trajectories was carried out using custom MATLAB scripts. For each single molecule, the intensity of the donor (Alexa Fluor 488), acceptor (Alexa Fluor 555) and corresponding FRET, along with the colocalized SWR1-binding intensity (Atto647N) were inspected. Nucleosomes that underwent exchange were identified by stepwise increases in the FRET trajectory. SWR1 binding was identified as an increase in the Atto647N intensity. Nucleosomes where the signal for SWR1 binding overlapped with at least one exchange event were included for further analysis. Dwell times were collected by manual inspection of the trajectories. Data were obtained by measuring several regions of interest from at least three independent slides. Dwell time plots were generated in MATLAB and plotted and fit in Igor Pro 8.
Single-molecule measurements of SWR1 nucleosome lifetime
Nucleosomes labelled with Alexa Fluor 488 on the short 2-bp overhang (113N2.AF488) were immobilized in a flow chamber as described above. Of SWR1(647N), 5ânM in imaging buffer (25âmM Tris-HCl (pH 7.8), 100âmM KCl, 4% glycerol, 1âmM EDTA, 2âmM MgCl2, 0.2âmgâmlâ1 BSA, Trolox, 2.5âmM protocatechuic acid and 0.25âµM protocatechuate-3,4-dioxygenase with 1âmM ATP) was injected. The three-colour smFRET microscope described above was used. The two excitation lasers (488ânm and 637ânm) were alternated at a frequency of 1âHz. Experiments were carried out at room temperature (22â°C). Trajectories in which SWR1(647N) colocalized with a nucleosome were selected and further processed using tMAVEN41 to determine the time for SWR1 to bind and the time SWR1 remained bound to a nucleosome.
Preparation of the SWR1ânucleosome complex for cryo-EM
Recombinant SWR1 was produced in BTI-TN-5B1-4 (High Five) insect cells, and the SWR1ânucleosome complex was assembled as previously described12. SWR1ânucleosome grids were prepared as previously described, except instead of glow discharge, the grids were cleaned by washing with water and ethyl acetate. Cryo-EM data acquisition, image acquisition and structure reconstruction were conducted using a similar procedure as previously described12. Data processing and refinement statistics for the two cryo-EM structures are summarized in Extended Data Table 1.
Cryo-EM data collection
A total of 35,076 micrographs were collected using a Titan KRIOS microscope operated at 300âkV. Images were collected on a Falcon IV direct electron detector with a pixel size of 1.1âà âpxâ1. Images were collected with a defocus range of â0.7 to â1.9âµm, with 1.0 s exposure time and a total dose of 40âeââà â2 fractionated over 39 frames.
Cryo-EM data processing
Movie frames were aligned using MotionCor2 (ref. 42), as previously described12. Contrast transfer function parameters were determined using Gctf43 as previously described12. Particle picking was performed in cryoSPARC44, as previously described12. Global-resolution and local-resolution estimates were calculated based on the gold-standard Fourier shell correlation (FSCâ=â0.143) criterion.
The cryo-EM processing workflow for the 3.8âà SWR1ânucleosome map in configuration I is summarized in Extended Data Fig. 6. First, in the recently collected SWR1ânucleosome dataset, 2D classification in cryoSPARC for 2D classes containing density for SWR1 or the nucleosome resulted in a working particle pool of 1,918,312 particles44. These were subdivided into three classes via heterogeneous refinement in cryoSPARC, resulting in class 1 (SWR1ânucleosome complex (15%)), class 2 (SWR1-apo (55%)) and class 3 (nucleosome only (30%)). The subset of 268,805 particles in class 1 (SWR1ânucleosome) was then further classified into five classes via heterogeneous refinement in cryoSPARC, resulting in class 1.1 (SWR1ânucleosome in configuration I (68%)), class 1.2 (SWR1ânucleosome configuration II (17%)), class 1.3 (poorly aligned class (9%)), class 1.4 (poorly aligned class (2%)) and class 1.5 (poorly aligned class (4%)). The particles in class 1.1 were then imported and subjected to 3D refinement in RELION before one round of 3D classification without alignment (Tâ=â30), with a soft mask overlapping the Swc2âbottom gyre DNA interface45. This generated two classes: class 1.1.1 (no density for bottom gyre DNA (63%)) and class 1.1.2 (clear density for bottom gyre DNA (37%)). Particles in class 1.1.2 were further selected for 3D refinement in RELION.
Next, in the previously collected dataset, 2D classification in cryoSPARC for 2D classes containing density for SWR1 or the nucleosome resulted in a working particle pool of 296,061 particles. These were subdivided into three classes via heterogeneous refinement in cryoSPARC, resulting in a class 1.1 (SWR1ânucleosome complex (33%)), class 1.2 (SWR1-apo (39%)) and class 1.3 (nucleosome only (28%))44. The subset of 96,648 SWR1ânucleosome particles were then further classified into five classes via heterogeneous refinement in cryoSPARC, resulting in class 1.1 (SWR1ânucleosome in configuration I (68%)), class 1.2 (SWR1ânucleosome configuration II (23%)), class 1.3 (poorly aligned class (5%)), class 1.4 (poorly aligned class (2%)) and class 1.5 (poorly aligned class (2%)). Particles in class 1.1 were imported and refined in RELION before one round of 3D classification without alignment (Tâ=â30), with a soft mask overlapping the Swc2âbottom gyre DNA interface. This generated two classes: class 1.1.1 (no density for bottom gyre DNA (16%)) and class 1.1.2 (clear density for bottom density (84%)). Particles in class 1.1.2 were further selected for 3D refinement in RELION45. Particles from classes 1.1.2 in the recently collected dataset and 1.1.2 in the previously collected dataset were then merged to generate a working pool of 123,591 particles. The resulting particles were then subjected to 3D refinement and contrast transfer function refinement in RELION with a mask corresponding to the SWR1 subcomplex of Swr1, Arp6, Swc6, Swc2, RuvBL1 and RuvBL2, and the nucleosome to generate the final 3.8âà SWR1ânucleosome map in configuration I45.
The cryo-EM processing workflow for the 4.7âà SWR1ânucleosome map in configuration II is summarized in Extended Data Fig. 6. First, in the recently collected SWR1ânucleosome dataset, particles in class 1.2 were selected, generating a working pool of 35,102 particles. The subset of particles was further classified into two classes in RELION using 3D classification with alignment (Tâ=â6) in the absence of a mask45. This generated class 1.2.1 (SWR1ânucleosome with poor density for the upper gyre DNA (39%)) and class 1.2.2 (SWR1ânucleosome with clearer density of upper gyre DNA (61%)). The particles in class 1.2.2 were selected, generating a working pool of 20,990 particles for 3D refinement in RELION.
Next, in the previously collected SWR1ânucleosome dataset, particles in class 1.2 were selected, generating a working pool of 21,054 particles. The subset of particles was further classified in two classes in RELION using 3D classification with alignment (Tâ=â6) in the absence of a mask45. This generated class 1.2.1 (SWR1ânucleosome with poor density for the upper gyre DNA (40%)) and class 1.2.2 (SWR1ânucleosome with clearer density of upper gyre DNA (60%)). The particles in class 1.2.2 were selected, generating a working pool of 12,605 particles for 3D refinement in RELION45. Particles from classes 1.2.2 in the recently collected dataset and 1.2.2 in the previously collected dataset were then merged to generate a working pool of 33,595 particles. The resulting particles were then subjected to 3D refinement and contrast transfer function refinement in RELION with a mask corresponding to the SWR1 subcomplex of Swr1, Arp6, Swc6, Swc2, RuvBL1 and RuvBL2, and the nucleosome to generate the final 4.7âà SWR1ânucleosome map in configuration II.
Model building
For the Swc2 subunit, an initial template was generated using AlphaFold25. Different regions corresponding to secondary structures of the template were manually truncated and docked separately into the recently generated 3.8âà SWR1ânucleosome map in configuration I in Chimera12,46, before being further built in Coot47. The final coordinates were subjected to real-space refinement in Phenix48.
For the 3.8âà SWR1ânucleosome configuration I map, first the SWR1ânucleosome complex from the previously solved 3.6âà SWR1ânucleosome structure (Protein Data Bank (PDB) ID 6GEJ) was docked into the density using Chimera12,46. The coordinates for the DNA were then omitted. Next, the SWR1ânucleosome complex from the previously solved 4.5âà SWR1ânucleosome structure (PDB ID 6GEN) was superimposed onto the docked structure using RuvBL1 and RuvBL2 as a reference. Coordinates for the superimposed structure were then omitted, with exception to the coordinates for the DNA, which was kept and docked into the 3.8âà SWR1ânucleosome configuration I map in Chimera, before merging the two PDB models: SWR1ânucleosome DNA omitted and DNA only together. The coordinates corresponding to the previously built Swc2 subunit were then omitted, and the coordinates for the newly built Swc2 model were docked into the map. Additional DNA overhang was then built manually in Coot12,46,47. The final coordinates were then subjected to real-space refinement in Phenix48.
For the 4.7âà SWR1ânucleosome configuration II map, SWR1 from the previously solved 3.6âà SWR1ânucleosome structure (PDB ID 6GEJ) was docked into the density using Chimera46. The coordinates corresponding to Swc2 were omitted, and the recently built Swc2 was docked together into the density using Chimera and further built in Coot46,47. The additional DNA overhang was then built manually in Coot. The final coordinates were then subjected to real-space refinement in Phenix48.
2D classification of SWR1-mediated nucleosome flipping
First, in the recently collected SWR1ânucleosome dataset, particles in class 2 (SWR1-apo (55%)) were selected, generating a working pool of 594,100 particles. The subset of particles was then further classified into four classes via heterogeneous refinement in cryoSPARC, resulting in class 2.1 (RuvBL1âRuvBL2 only (21%)), class 2.2 (a poorly aligned class (20%)), class 2.3 (SWR1-apo with additional density underneath SWR1 (38%)) and class 2.4 (a poorly aligned class (21%)). Particles in class 2.3 were then selected for 2D classification in RELION45.
Next, in the previously collected SWR1ânucleosome dataset, particles in class 2 (SWR1-apo (39%)) were selected, generating a working pool of 115,463 particles. The subset of particles was then further classified into four classes via heterogeneous refinement in cryoSPARC, resulting in class 2.1 (RuvBL1âRuvBL2 only (25%)), class 2.2 (a poorly aligned class (20%)), class 2.3 (SWR1-apo with additional density underneath SWR1 (30%)) and class 2.4 (a poorly aligned class (25%)). Particles in class 2.3 were then selected for 2D classification in RELION. The particles in class 2.3 in the recently collected SWR1ânucleosome dataset and the particles in class 2.3 in the previously collected SWR1ânucleosome dataset were then merged and subjected to multiple rounds of 2D classification in RELION to obtain 2D classes of SWR1-mediated nucleosome flipping.
Statistics and reproducibility
For data relating to Fig. 1, the total number of traces used in each dataset is indicated in each panel and was derived from three independent experiments. For data relating to Fig. 2, the total number of traces used for each dataset is indicated in each panel and was derived from four independent experiments. For data relating to Figs. 3 and 4, two independent experiments were performed, one of which is shown. The total number of traces used for each dataset is indicated in each panel. All gels were independently and successfully repeated twice.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.