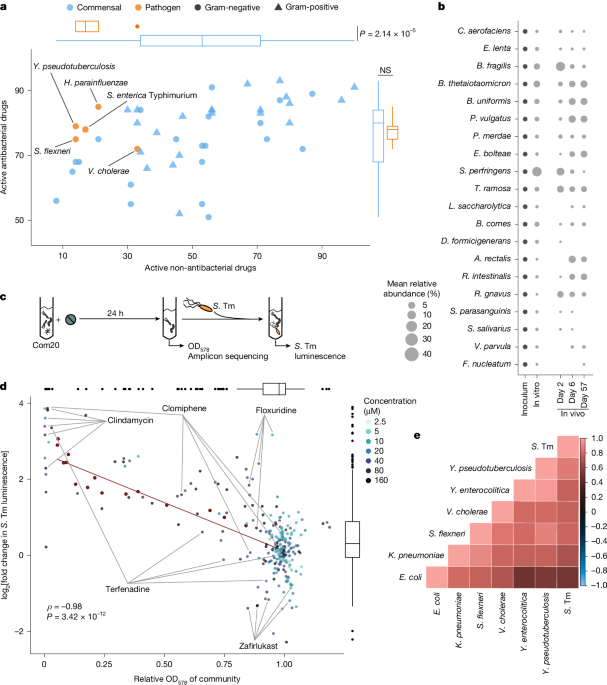
Bacterial cultivation of monocultures, Com20, Com21 and stool-derived communities
The species used in this study are listed in Supplementary Table 13. They were purchased from the Leibniz-Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, BEI Resources, the American Type Culture Collection or Dupont Health & Nutrition, or were provided as gifts from the Denamur Laboratory (INSERM), the Blokesch Laboratory (EPFL), the Andrews–Polymenis Laboratory (Texas A&M University), the Darby Laboratory (UCSF) or the Wagner Laboratory (University of Tübingen). All gut commensal species, whether grown individually or as a community, were cultivated in mGAM (Nisui Pharma Solutions) at 37 °C, with the exception of Veillonella parvula and Bilophila wadsworthia monocultures. We cultured V. parvula in Todd–Hewitt broth supplemented with 0.6% sodium lactate and B. wadsworthia in mGAM supplemented with 60 mM sodium formate and 10 mM taurine. We pre-reduced the medium for a minimum of 24 h under anoxic conditions (2% H2, 12% CO2 and 86% N2) in an anaerobic chamber (Coy Laboratory Products). The species were inoculated from frozen stocks into liquid culture medium and passaged twice (1:100) overnight to ensure robust growth. We periodically verified the purity and identity of the species through sequencing of the 16S rRNA gene and/or MALDI–TOF mass spectrometry38.
We selected a set of 31 prevalent and abundant species from the human gut microbiome, which differed by >3% in their 16S rRNA gene sequences in the V4 region. When monocultures of all species were mixed in equal OD ratios, 20 of these species were consistently detectable and their levels were stable after several passages15.
For experiments involving human stool-derived material, informed consent was obtained from all eight donors (approved by the Ethics Committee of the University Hospital Tübingen, project ID 314/2022B02). We generated stool-derived communities from fresh human faecal samples as previously described39,40 by inoculating from frozen glycerol stocks into 3 ml BHI and serial dilution of 1:200 for three 48-h passages. This process ensured that the composition reached a steady state before measurements. We performed the experiments in clear, flat-bottomed 96-well plates (Greiner Bio-One). We sealed the plates with breathable AeraSeals (Excel Scientific). Communities were stored with glycerol as frozen stocks at −80 °C, inoculated from frozen stocks into fresh mGAM and grown overnight.
We carried out selective plating of pathogens under aerobic conditions. For animal experiments, we cultured S. Tm in LB broth supplemented with 0.3 M NaCl and determined S. Tm loads in intestinal content and organs on MacConkey agar supplemented with 50 µg ml–1 streptomycin.
Prestwick library screening for pathogens
We carried out the Prestwick library screening as previously described1 on five pathogenic bacterial species in mGAM under anaerobic conditions. In brief, the library, which consists of 1,197 drugs approved by the Food and Drug Administration, was diluted to 100-fold the working concentration in DMSO (2 mM) in V-bottom polypropylene plates (Greiner Bio-One, 651261). For the screening experiments, we diluted drug master plates to 2-fold the working concentration in mGAM (40 µM) in U-bottom plates (Thermo Scientific, 168136), aliquoted them (50 µl per plate) and stored them at −20 °C for a maximum of 1 month. The DMSO control wells in each 96-well plate served as controls. For experiments with H. parainfluenzae, we supplemented mGAM with 0.5 mg l–1 hemin and 2 mg l–1 NAD. Before inoculation, we pre-reduced the drug plates overnight in an anaerobic chamber.
Before the screening experiments, we passaged bacterial strains twice overnight (1:100) anaerobically and adjusted the OD578 to 0.02. After inoculation, the starting OD578 for all bacterial species was 0.01 and the drug concentration in the plate was 20 μM with 1% DMSO. We sealed all plates with breathable membranes (Breathe-Easy, Sigma-Aldrich, Z380059). Bacterial growth was tracked by measuring the OD578 every hour for 24 h using a microplate spectrophotometer (EON, Biotek) coupled with a Biostack 4 microplate stacker (Biotek), both housed inside an incubator (EMBL workshop). All screening experiments were performed in three biological replicates. For analysis, we truncated growth curves at the transition from the exponential to the stationary phase for analysis. We then calculated the area under the curve (AUC) using the trapezoidal rule and normalized it to the solvent or DMSO controls in the same plate. We identified hits from normalized AUC measurements by fitting heavy-tailed distributions, specifically the scaled Student’s t-distribution41, to the wells containing controls. We combined P values for each drug and strain across replicates using Fisher’s method, and calculated the false-discovery rate using the Benjamini–Hochberg method over the entire matrix.
To compare drug effects between P. vulgatus WT and P. vulgatus ΔBVU_1672-1675, we used an updated version of the Prestwick library, which contains 1,520 drugs. We performed the screen as described for the pathogens and calculated the normalized median AUC per drug–strain combination1,21. We defined compounds that reduced the median AUC below 0.1 as hit compounds and compared total hit counts and counts by class (antibiotic or non-antibiotic drug).
Targeted gene deletions in P.
vulgatus
We generated genomic knockouts of target genes in P. vulgatus as described elsewhere42. In brief, the method we used relies on a two-step allelic exchange by homologous recombination. For this, the regions flanking the gene of interest 1,500 bp upstream and downstream were amplified from genomic DNA and cloned into the linearized anhydrotetracycline (aTc)-inducible suicide vector pLGB13 using HiFi–Gibson assembly. This plasmid contains an ampicillin-resistance cassette for maintenance in E. coli, an erythromycin-resistance cassette as a selection marker in Bacteroidota species and the aTc-inducible ssBfe1 counter selection cassette, which will express the highly toxic effector Bfe1 from the type VI secretion system of Bacteroides fragilis43. We transformed the vector that contained the flanking regions into E. coli DATC44 using heat-shock transformation. We then conjugated the plasmid into P. vulgatus, where it integrates into the chromosome at the site of the gene of interest through homologous recombination under erythromycin selection. Using aTc counterselection, which induces ssBfe1-mediated rapid cell death, colonies that underwent a second homologous recombination, thereby losing the integrated plasmid again, were selected. We analysed colonies by PCR and Sanger sequencing to verify whether they were WT revertants or knockouts.
Targeted gene deletions in S. Tm
We introduced ΔfrdD::aphT into S. Tm SB300 from S. Tm 14028 ΔfrdD::aphT45 through P22 phage transduction and subsequent selection on kanamycin. Successful phage transduction was confirmed by PCR using tag-specific primers (aphT_fwd: 5′-CTGGCTGCTATTGGGCGAAG-3′; frdD_rev: 5′-GATTCACATCTTGGACCGCC-3′).
Drug selection
We selected drugs for the in vitro challenge assay on the basis of their direct inhibitory effect on members of Com20 in monocultures1. We aimed to identify drugs with different inhibition profiles across the 20 species so that we could generate communities with sufficient compositional variation. We performed hierarchical clustering (Euclidean distance metric and complete linkage method) using the normalized AUC values of the 172 drugs that showed significant inhibition (adjusted P < 0.01) against at least 5 of the 20 species in Com20. From these clusters, we selected 63 drugs that represented diverse inhibition spectra across Com20 members (Extended Data Fig. 1a). We also included 3 drugs not part of the Prestwick library, which resulted in a panel of 65 drugs for the initial assays in Com20 members. This set was further streamlined as we progressed through the experiments, as illustrated in Extended Data Fig. 1a. None of the drugs interfered with the luminescence readout of the assay. We excluded drugs that directly inhibited S. Tm growth with IC25 values < 5 µM (nalidixic acid and norfloxacin; Supplementary Table 2). We further excluded β-lactam antibiotics owing to the presence of ampicillin resistance on the pilux luminescence plasmid used in the S. Tm invasion assay.
IC25 determination
We dissolved all drugs in DMSO, except for clomipramin, doxorubicin and tobramycin, which were dissolved in water. We prepared drug master plates at a concentration 100 times the working concentration by serially diluting the stock solutions 2-fold in DMSO or water. We diluted column-wise in V-bottom 96-well plates (Greiner Bio-One, 651261), starting from 160 mM. Each column in the plate contained eight twofold dilutions of a drug, except for column 7, which contained DMSO or water as a control. This strategy resulted in 11 drugs screened per plate. We diluted the plates to 2 times the assay concentration in 50 µl mGAM in U-bottom 96-well plates (Thermo Scientific, 168136) and stored them at −20 °C for a maximum of 1 month. Before the assay, we thawed and pre-reduced the plates overnight in an anaerobic chamber.
Monocultures or stool-derived communities21 were grown overnight in 5 ml mGAM. The next day, we diluted the communities to an OD578 of 0.02. We then added 50 μl of the suspension to the drug plates to result in a starting OD578 of 0.01 and a DMSO concentration of 1% in all wells. We sealed plates with Breathe-Easy membranes (Sigma-Aldrich, Z380059). Growth curves at OD578 were monitored every hour after 1 min of linear shaking under anaerobic conditions using an Epoch2 microplate reader coupled with a Biostack 4 microplate stacker (both Agilent) housed in a custom-made incubator (EMBL workshop21). We analysed at least three biological replicates for each species.
To calculate the AUC, we used the R package neckaR (https://github.com/Lisa-Maier-Lab/neckaR), using control wells in the plate that did not contain any drugs to define normal growth. We calculated the median AUC for each concentration across the three replicates. To conservatively remove the effects of noise, we enforced monotonicity. If the AUC decreased at lower concentrations, it was set to the highest AUC measured at higher concentrations. The IC25 was defined as the lowest concentration at which a median AUC < 0.75 was observed.
Assessment of correlations between phylogenetic relatedness and responses to bacterial and non-antibiotic drugs
To evaluate the association between the response of microorganisms to antibiotics and non-antibiotics in an evolutionary context, we used the newly obtained AUCs from the Prestwick library screen, together with those of previously reported commensal bacteria1, to determine the similarity of the response between bacterial species, as measured by the Euclidean distance. Only drugs that inhibited the growth of >5 species were included in the analysis. We used the R package ape (v.5.8)46 to calculate the cophenetic distances between species; the phylogeny was reconstructed using a multilocus alignment obtained from whole bacterial genomes using phylophlan (v.3.0)47. Principal coordinate plots were generated from Euclidean distance matrices. We tested the global association between AUC and phylogenetic distances using the Mantel test as implemented in the R package Ade4 (v.1.7-22)48. We further assessed the change in the strength of the association across evolutionary distances using a phylocorrelogram with the R package phylosignal (v.1.3.1)49, which shows the correlation between the overall response to the drugs and the phylogenetic distance between species as a function of the phylogenetic distance.
Prediction of antimicrobial resistance and stress-related genes in pathogens and gut commensals
To assess the genetic repertoire of antimicrobial resistance and stress response genes in gut commensals and pathogens, we first performed gene calling from whole genome sequences using FragGeneScan (v.1.31)50. We identified genes involved in antimicrobial resistance, efflux and stress responses using AMRFinderPlus (v.3.11)51 and the database (v.2023-09-26.1) with amino acid sequences as input. We then used argNorm (v.0.2)52 to normalize the antibiotic-resistance gene annotations.
Preparation of drug master plates for in vitro invasion assays for S. Tm
For each drug, we tested five concentrations. Drugs with reported intestinal concentrations exceeding 20 µM (ref. 1) were screened at concentrations from 10 to 160 µM, and the remaining drugs were screened at concentrations from 2.5 to 40 µM.
We prepared master plates in V-bottom 96-well plates at 100-fold the drug working concentration in DMSO as described above. Concentration gradients of the drugs (16 mM, 8 mM, 4 mM, 2 mM and 1 mM or 4 mM, 2 mM, 1 mM, 0.5 mM and 0.25 mM) were represented by each column, with rows B and G having the highest and lowest concentration, respectively. In each deep-well plate, row E served as solvent controls that contained only DMSO or water. To prepare the 96-deep-well plates (Thermo Fisher Scientific, AB-0564) for the S. Tm challenge assay, we transferred 5 µl of the drug master plate to the deep-well plate, which already contained 95 µl mGAM. Subsequently, we pre-reduced the deep-well plates overnight (5 times the drug working concentration in 5% DMSO) in an anaerobic chamber. Wells on the border contained only mGAM (sterile controls).
Assembly of Com20 and Com21 for in vitro invasion assays for S. Tm
For Com20 and Com21 assembly, we inoculated each member from frozen stocks and cultured them anaerobically in 5 ml mGAM over two overnight passages (1:100) as monocultures. We measured the OD578 individually for each species. We mixed together the cultures in the volume required to achieve a total OD578 of 0.0125 (for example, in Com20, each species contributed an equal OD578 of 0.000625) and 400 µl of this suspension was added to wells of 96-deep-well plates that contained drugs as described above to achieve a starting OD578 of 0.01 (total volume of 500 µl).
We sealed the deep-well plate with the drugs and communities with a Breathe-Easier membrane (Sigma-Aldrich, Z763624) and incubated them anaerobically at 37 °C for 24 h. The 24 h of incubation with drugs disrupted the composition of the communities, and we used the disrupted communities for the in vitro S. Tm challenge assay. We obtained pellets from 300 µl of the cultures, which we then froze for 16S rRNA gene analysis.
In vitro S. Tm challenge assay
For the luminescence-based invasion assay, we used the human gut pathogen S. Tm strain SB300 (ref. 53) with the plasmid pIJ11282 ilux (pRS16591, S. Tm pilux; a gift from the Foster Laboratory, University of Oxford) for constitutive expression of the ilux operon under the nptII promoter54. S. Tm pilux was grown anaerobically at 37 °C overnight in mGAM supplemented with 100 µg ml–1 ampicillin and then subcultured by diluting 1:100 in the same medium. The next day, we measured the OD578 of 100 µl of all drug-perturbed communities in a 96-well clear, flat-bottom plate. To assess the growth potential of S. Tm pilux in the drug-perturbed communities, we transferred 50 µl from each well of the drug-perturbed communities into new pre-reduced, deep-well plates. We diluted S. Tm pilux to an OD578 of 0.0025 and added 200 µl of this suspension to the assay deep-well plate. We added 250 µl mGAM so that the total volume was 500 µl, which resulted in a starting OD578 for S. Tm of 0.001 and for the untreated community of 0.5. Of note, this protocol resulted in the transfer of residual amounts of the drug, up to 10% of the original concentration. We sealed the assay plate with a Breathe-Easier membrane (Sigma-Aldrich, Z763624) and incubated it anaerobically at 37 °C for 4.5 h. Thereafter, the plate was taken out of the anaerobic chamber. We thoroughly mixed the contents of the wells and added 25 µl of 2 mg ml–1 chloramphenicol to each well to halt S. Tm growth and to stabilize the luminescence signal. The cell suspension (100 µl) was transferred to a white 96-well plate (Thermo Fisher, 236105). Approximately 10 min later, the plate was incubated for 10 min at 37 °C in a Tecan Infinite 200 PRO microplate reader and luminescence was measured.
We obtained two measurements: the OD578 of communities after overnight incubation with the drugs, and the luminescence emitted by S. Tm as a proxy for pathogen growth in the drug-perturbed communities. For data analysis, OD578 values were first corrected by subtracting the baseline OD578 from mGAM. Then, we normalized the luminescence and OD578 values to the control column in row E, which contained the unperturbed community (solvent controls) by dividing values of perturbed communities by values of untreated communities. Both S. Tm luminescence and Com20 OD578 were highly correlated among the three replicates (R2 = 0.56–0.74 and R2 = 0.85–0.9, respectively; Extended Data Fig. 4e).
Post-wash S. Tm challenge assay
We evaluated whether washing drug residue from the community after treatment affected S. Tm growth. The post-wash S. Tm challenge assay was conducted following the same protocol as the S. Tm challenge assay, with the modification that the procedure was carried out in 1.5 ml Eppendorf tubes. We tested the following drugs: 20 µM clotrimazole, 80 µM zafirlukast, 160 µM chlorpromazine and 80 µM terfenadine from the colonization group S. Tm neutral; 80 µM clomiphene, 20 µM floxuridine, 20 µM erythromycin and 80 µM sertindole from the colonization group S. Tm favouring; and 1% DMSO as a control. Each condition was carried out in two Eppendorf tubes. After 24 h of drug treatment, we centrifuged one tube of each condition for 5 min at 3,000 g at room temperature under anaerobic conditions. The supernatant was removed and the pellet was resuspended in 500 µl mGAM. Next, we transferred 50 µl of each tube of each condition in triplicate to a deep-well plate containing 250 µl mGAM to compare the washed and the unwashed culture. Finally, S. Tm was added and the luminescence was measured after 4.5 h. Signals were normalized to S. Tm growing in the DMSO-treated community.
Pairwise co-culture and single-species dropout assays
We conducted pairwise co-culture assays to measure the contribution of each member of Com20 to S. Tm growth individually. The commensal species and S. Tm pilux were grown anaerobically overnight in mGAM and subcultured once before the experiment. On the following day, we mixed the commensal and S. Tm pilux in 96-deep-well plates with a total volume of 500 µl mGAM. We set the initial OD578 of the commensal to 0.1, whereas S. Tm had an initial OD578 of 0.0002 (commensal to pathogen ratio of 500:1), as described above for the S. Tm challenge assay in Com20. Control wells contained only S. Tm in monoculture. After a growth period of 4.5 h at 37 °C under anaerobic conditions, we calculated S. Tm levels as described for the in vitro S. Tm challenge assay.
The single-species dropout assay was performed in a similar way as for the in vitro S. Tm challenge assays. We assembled 19-member communities by omitting one strain at a time in the volume required to achieve a total OD578 of 0.5. Then, we mixed 800 µl of this suspension with 200 µl 50% glycerol (with a few crystals of palladium black (Sigma-Aldrich)). Communities preserved in 1.8 ml cryovials (Thermo Scientific NUNC, 10674511) were frozen at −80 °C and grown overnight twice anaerobically at 37 °C before conducting the pathogen challenge assay. We normalized the growth of S. Tm on dropout communities to the growth of the pathogen in Com20.
In vitro Transwell S. Tm challenge assay
To analyse the transcriptional profile of both the community and S. Tm after drug treatment, we performed the S. Tm challenge assay in a Transwell format. For this, we selected treatments that led to a low biomass of Com20 (160 µM terfenadine and 80 µM clomiphene) and drugs that affected community composition but not biomass (20 µM floxuridine and 80 µM simvastatin). Com20 and S. Tm were inoculated from a cryostock in 5 ml mGAM and incubated anaerobically overnight at 37 °C. We diluted the drugs and the control solvent (DMSO) in mGAM in glass tubes in a total volume of 4.5 ml. After overnight incubation, we measured the OD578 of Com20. Then, 500 µl Com20 at an OD578 of 0.05 was added to tubes containing mGAM and drug or solvent, which resulted in a total volume of 5 ml. This setup ensured a final drug concentration of 1× and an initial Com20 OD578 of 0.005. We incubated drug-treated Com20 anaerobically for 24 h at 37 °C; S. Tm was subcultured. The next day, we transferred Com20 and S. Tm to 6-well cell culture plates (Greiner, 657160) in a total volume of 7 ml per well. We added 2.3 ml mGAM to all wells and transferred 700 µl of each drug-treated community in triplicate to the 6-well cell culture plate. We placed the 6-well cell culture insert (CellQART, 0.4 µm, 9300402) in each well and filled all inserts with 4 ml S. Tm with an initial OD578 of 0.001. Plates were anaerobically incubated at 37 °C. After 4.5 h, we combined the 3 technical replicates in one 15 ml Falcon tube of either S. Tm or the drug-treated community to reach a minimum of 109 cells per tube. We took out the tubes from the anaerobic chamber to centrifuge them at 4,300g for 20 min at 4 °C. After centrifugation, we placed the tubes immediately on ice, removed the supernatant and added 1 ml TRIzol (Invitrogen by Thermo Fisher Scientific, 15596026). After vortexing the tubes, we transferred their content to a 2 ml Eppendorf tube before leaving the pellet for 10 min at room temperature and freezing it at −80 °C. We repeated the experiment three times across three different weeks.
Samples were sent to Novogene for RNA isolation and sequencing. In brief, total RNA was extracted using an in-house RNA purification kit and ribosomal RNA was removed using a Ribo-Zero Plus rRNA depletion kit (Illumina) followed by ethanol precipitation. After fragmentation, the first-strand cDNA was synthesized using random hexamer primers. During the second-strand cDNA synthesis, dUTPs were replaced with dTTPs in the reaction buffer. Directional libraries were generated using a Novogene NGS Stranded RNA Library Prep Set, which involved end repair, A-tailing, adapter ligation, size selection, USER enzyme digestion (New England Biolabs), amplification and purification. Libraries were sequenced using an Illumina Novaseq X Plus-PE150 platform.
Quantification of S. Tm in treatment-mimicking communities
To validate our screen, we selected four conditions: treatment with erythromycin, floxuridine, sertindole or zafirlukast. On the basis of the composition of Com20 after treatment with these drugs, we assembled treatment-mimicking communities that contained only the members with a mean relative abundance of ≥3% after 24 h of drug exposure. We incubated Com20 and treatment-mimicking communities in deep-well plates at 37 °C anaerobically. After 24 h, we performed a dilution series of these communities in deep-well plates in a total volume of 400 µl. We transferred 100 µl of each dilution and Com20 to flat-bottom plates and measured the OD578. Fifty microlitres was transferred to new deep-well plates containing 250 µl mGAM per well. In addition, 200 µl S. Tm pilux (OD578 0.0025) was added to each well and the plate was incubated for 4.5 h at 37 °C. We retained the remaining volume of the dilution series for DNA isolation and 16S rRNA gene sequencing. After 4.5 h, we measured S. Tm luminescence as described above. We performed the experiment in triplicate, and the luminescence measurements in treatment-mimicking communities were normalized to the luminescence in Com20. We compared the log2[fold change] of S. Tm luminescence and the OD578 between treatment-mimicking communities and drug-treated communities to identify the dilution step that best matched the drug-treated community.
S. Tm WT and S. Tm ΔfrdD::aphT competition assay
To test whether fumarate respiration has an essential role in outcompeting a close niche competitor, we performed a competition experiment between E. coli ED1α and Salmonella. E. coli ED1α, S. Tm SB300 WT, S. Tm SB300 ΔfrdD::aphT and S. Tm SB300 WITS-tag (kanamycin resistant55), which were inoculated anaerobically for 2 nights at 37 °C. Next, we measured the OD578 of all bacteria and mixed them in a total volume of 5 ml. E. coli ED1α was added to every condition with an initial OD578 of 0.5. In condition one, we tested the growth of Salmonella by adding S. Tm WT and S. Tm WITS-tag with an initial OD578 of 0.0005. In condition two, we tested the growth of S. Tm lacking the subunit D of fumarate reductase by adding S. Tm WT and S. Tm ΔfrdD::aphT with an initial OD578 of 0.0005. We incubated tubes at 37 °C anaerobically for 24 h before plating out on selective agar. Condition one was plated out on LB agar with streptomycin (50 µg ml–1; from herein on LBStrep) to obtain all Salmonella counts and on LB agar, streptomycin (50 µg ml–1) and kanamycin (30 µg ml–1, henceforth LBStrepKan) to obtain counts from S. Tm WITS-tag. Condition two was plated out on the same selective agar plates; although LBStrep was used to obtain all Salmonella counts, on LBStrepKan, only S. Tm ΔfrdD::aphT was able to grow. We subtracted LBStrepKan counts from LBStrep counts to obtain S. Tm WT and calculated their ratio. The experiment was performed in six replicates.
To verify our findings in a community context, we selected streptozotocin (40 µM), which targets E. coli ED1α, tiratricol (160 µM), which does not affect E. coli ED1α, and DMSO as a control solvent. We used Com20, which served as a simple synthetic community, Com21, because it mirrors Com20 but contains a niche competitor (E. coli ED1α) and the stool-derived community, because of its complexity. All three communities were inoculated from cryogenic stock in 5 ml mGAM anaerobically and incubated for one night at 37 °C. S. Tm WT and S. Tm ΔfrdD::aphT were inoculated from plates in 5 ml mGAM anaerobically and incubated for one night at 37 °C. We added drugs and control solvent (DMSO) to a total amount of 4.5 ml mGAM in glass tubes. The next day, the OD578 of all communities was determined, and communities were diluted to an initial OD of 0.005 and added to tubes containing mGAM and drugs or solvent in a total volume of 5 ml, which resulted in a 1× drug concentration. Drug-treated communities were incubated at 37 °C for 24 h anaerobically and Salmonella was subcultured. After 24 h, drug-treated communities were 10× diluted into fresh mGAM and the OD578 of S. Tm was determined. We added S. Tm WT and S. Tm ΔfrdD::aphT to each drug-treated community with an initial OD of 0.0005 and incubated these tubes for 24 h at 37 °C anaerobically. The following day, we plated each condition on either LBStrep to obtain all S. Tm counts or on LBStrepKan to count only S. Tm ΔfrdD::aphT. We subtracted S. Tm ΔfrdD::aphT counts from all S. Tm counts to obtain S. Tm WT counts and calculated the ratio of S. Tm WT/S. Tm ΔfrdD::aphT. The experiment was repeated 5–6 times.
Plasmid transformation of pathogenic Enterobacteriaceae
We incubated bacterial strains overnight in 6 ml LB medium at 27 °C (WA-314, YpsIII) or 37 °C (Kp MKP103, Ec CFT073). We centrifuged the overnight cultures for 5 min at 4,000g, washed the pellets twice with 5 ml of 300 mM sucrose solution, transferred 1 ml of 300 mM sucrose solution to an Eppendorf cap and centrifuged for 1 min at 10,000g. The supernatant was removed, the bacteria were resuspended in 100 µl of 300 mM sucrose solution and transferred to a Gene Pulser cuvette (0.2-cm electrode gap, Bio-Rad), and 100 ng of plasmid DNA (pEB1GM or pEB2GO, synthesized by GenScript) was added. Subsequently, we carried out electroporation using a Gene Pulser (Bio-Rad) and immediately added 1 ml LB. The bacterial suspension was then shaken at the corresponding temperature for 1 h and plated on LB gentamicin plates (15 µg ml–1 for WA-314, YpsIII and Kp Ec CFT073; 75 µg ml–1 for Kp MKP103) overnight. We verified the success of the electroporation by measuring chemiluminescence of the lux reporter.
Adaptation of the S. Tm challenge assay to other pathogens
We screened other Gammaproteobacteria species in a similar manner to S. Tm in the challenge assay described above. We prepared drug master plates in the same way, except that we tested 10 drugs and the master plate concentration ranged from 10 mM to 1 mM. Moreover, only the outer rows were left empty to serve as medium controls. We tested post-treatment expansion of Gammaproteobacteria species in Com20, which we assembled as described above. For the luminescence-based assay, we used the human gut pathogens E. coli CFT073, K. pneumoniae MKP103, S. flexneri 24570, Y. enterocolitica WA-314, Y. pseudotuberculosis YPIII and V. cholerae A1552. With the exception of V. cholerae, all pathogens contained a variant of the pilux plasmid that enabled constitutive expression of the lux reporter. We incubated all pathogens anaerobically overnight at 37 °C in mGAM supplemented with 100 µg ml–1 ampicillin (S. flexneri), 15 µg ml–1 gentamicin (E. coli, Y. enterocolitica and Y. pseudotuberculosis) or 75 µg ml–1 gentamicin (K. pneumoniae) and then subcultured by diluting 1:100 in the same medium. We proceeded as for the in vitro S. Tm challenge assay but incubated the plates at 37 °C for a species-specific amount of time (4.5 h for E. coli, 5 h for S. flexneri and K. pneumoniae, 5.5 h for V. cholerae and 7 h for Y. enterocolitica and Y. pseudotuberculosis).
To measure the growth of V. cholerae, we serially diluted the plates (101–108-fold) in PBS and selectively plated aerobically on LB agar with 100 µg ml–1 ampicillin for pathogen enumeration. For the other pathogens, we measured their growth as described for S. Tm pilux with a Tecan Infinite 200 PRO microplate reader. For each treatment, we obtained two measurements: the OD578 of communities after overnight incubation with the drugs and the luminescence emitted by the pathogens (CFU in the case of V. cholerae) as a proxy for pathogen growth in the drug-perturbed communities. For data analyses, both the luminescence (CFU for V. cholerae) and OD578 values were normalized to the median of the controls in row E, which contained the unperturbed community (solvent controls).
We did not evaluate the effect of washing the community after drug treatment but before pathogen introduction given the results we obtained on a similar experiment using S. Tm (see the section ‘Post-wash S. Tm challenge assay’). Moreover, even when the IC25 of the pathogens was low, such as in the case of floxuridine (Supplementary Table 2), community treatment at the highest concentrations of the compound led to an increased pathogen growth (Supplementary Table 5), despite the potentially disrupting effect of the residual drug.
General statistical analyses
We used R (v.4.2.0) for data processing and formatting. The package ggplot2 (v.3.5.1) was used for visualization. For hypothesis testing, t-tests, Kruskal–Wallis tests and Wilcoxon tests were performed as implemented in the package Rstatix (v.0.7.2).
Analysis of community composition using 16S rRNA gene amplicon sequencing
DNA was extracted from pellets of 300 μl culture using a DNeasy UltraClean 96 Microbial kit (Qiagen, 10196-4) or from whole faecal pellets using a DNeasy PowerSoil HTP 96 kit (Qiagen, 12955–4). Library preparation and sequencing was performed at the NGS Competence Center NCCT. Genomic DNA was quantified with a Qubit dsDNA BR/HS Assay kit (Thermo Fisher) and adjusted to 100 ng input for library preparation. The first step PCR was performed in 25 µl reactions that included KAPA HiFi HotStart ReadyMix (Roche), 515F56 and 806R57 primers (covering about 350-bp fragment of the 16S V4 region) and template DNA (PCR program: 95 °C for 3 min, 28× (98 °C for 20 s, 55 °C for 15 s, 72 °C for 15 s), 72 °C for 5 min). Initial PCR products were purified using 28 µl AMPure XP beads and eluted in 50 µl of 10 mM Tris-HCl. Indexing was performed in a second step PCR that included KAPA HiFi HotStart ReadyMix (Roche), index primer mix (IDT for Illumina DNA/RNA UD Indexes, Tagmentation) and purified initial PCR product as template (PCR program: 95 °C for 3 min, 8× (95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s), 72 °C for 5 min). After another round of bead purification (20 µl AMPure XP beads, eluted in 30 µl of 10 mM Tris-HCl), the libraries were checked for correct fragment length on an E-Base device using E-Gel 96 Gels with 2% mSYBR Safe DNA gel stain (Fisher Scientific), quantified with a QuantiFluor dsDNA system (Promega) and pooled equimolarly. The final pool was set to 4 nM (Illumina standard value) before being brought to a loading concentration of 8 pM. The pool was sequenced on an Illumina MiSeq device with a v.2 sequencing kit (input molarity 10 pM, 20% PhiX spike-in, 2 × 250 bp read lengths).
Computational processing of 16S rRNA amplicon sequences
We used the R package DADA2 (v.1.21.0)58 following its standard operating procedure available from GitHub (https://benjjneb.github.io/dada2/bigdata.html). In brief, after inspecting the quality profiles of the raw sequences, we trimmed and filtered the paired-end reads using the following parameters: trimLeft: 23, 24; truncLen: 240, 200; maxEE: 2, 2; truncQ: 11. The filtered forward and reverse reads were de-replicated separately and used for inference of amplicon sequence variants (ASVs) using default parameters, after which the reads were merged on a per-sample basis. Next, we filtered the merged reads to retain only those with a length between 250 and 256 bp and carried out chimera removal.
We performed the taxonomic assignment in two steps. First, the final set of ASVs was classified up to the genus level using a curated DADA2-formatted database based on the genome taxonomy database (GTDB)59 (release R06-RS202; available at https://scilifelab.figshare.com/articles/dataset/SBDI_Sativa_curated_16S_GTDB_database/14869077). Next, ASVs belonging to genera expected to be in Com20 were further classified at the species level using a modified version of the aforementioned database that contained only full-length 16S rRNA sequences of the 20 members of the synthetic community. The sequence of each ASV was aligned against this database using the R package DECIPHER (v.2.24.0)60; we classified an ASV as a given species if it had sequence similarity of >98% to the closest member in the database. The abundance of each taxon of Com20 was obtained by aggregating reads at the species level. ASVs from in vitro communities and gnotobiotic mice were classified using the two-step processes; ASVs from SPF mice were classified using only the first step. We removed potential contaminant sequences from SPF mouse samples using the permutation filtering method implemented in the R package PERFect (v.1.14.0)61.
Overlap of pathways encoded by Com20 and Com21 and human gut metagenomes
We used the 16S rRNA gene abundance from control Com20 and Com21 in vitro communities and untreated gnotobiotic mice samples to predict the metabolic potential of the microbial communities using PICRUSt2 (v.2.4.1)62. As the composition of the synthetic communities is known, we retrieved the full-length sequences of the 16S rRNA gene for each of the member species and used them together with the species abundance data to predict metagenome functions. For our analyses, we used MetaCyc pathway abundances. We compared the number of metabolic pathways detected in the untreated in vitro and in vivo synthetic communities to actual human gut metagenomes. For this, we retrieved publicly available tables of MetaCyc pathway abundances processed using HUMAnN2 from GitHub (https://github.com/gavinmdouglas/picrust2_manuscript/tree/master/data/mgs_validation). These tables comprised 156 samples from the Human Microbiome Project63 and 57 samples from Cameroon64. For each set of samples, we considered a pathway as present if it was detected in ≥20% of samples.
Classification of drug treatments according to S. Tm growth
We grouped treatments according to their effect on Com20 in vitro. To do so, we calculated the mean normalized luminescence and the 95% confidence interval (CI) of each drug–concentration combination. A treatment was classified as S. Tm favouring if its mean normalized luminescence was >2 and the 95% CI did not span 2, whereas the treatment was classified as S. Tm restricting if the mean normalized luminescence was <0.5 and the 95% CI did not span 0.5. The treatment was classified as S. Tm neutral if the mean normalized luminescence was between 0.5 and 2. Communities with a normalized OD578 < 0.2 were also classified but marked for removal in downstream analyses to minimize the bias introduced by low-biomass samples.
Assessment of drug treatment on the composition of synthetic communities in vitro
To assess changes in microbial diversity after drug treatment, we calculated the species richness and Shannon’s index using the R package vegan (v.2.6-8)65. Tables of taxa abundances were rarefied to an even sequencing depth. We evaluated the association between S. Tm luminescence, OD578 of Com20 and these diversity measures by fitting linear models, with luminescence as the response variable. Our linear models evaluated each response variable separately or included OD578 and one diversity measure as the main effects. We estimated the proportion of variance explained by the models using the adjusted coefficient of determination (R2) and assessed the goodness-of-fit of each model with the Akaike information criterion.
We assessed differences in multivariate homogeneity of group dispersions between colonization groups (that is, S. Tm favouring, S. Tm restricting and S. Tm neutral) and untreated controls, and calculated differences in beta diversity with a PERMANOVA test on Bray–Curtis distance matrices using vegan (v.2.6-8). We performed pairwise PERMANOVA tests contrasting each treatment group to untreated controls accounting for normalized OD578 in the models; the OD578 value of control samples was set to 1. P values were adjusted using the Benjamini–Hochberg method and a significance threshold of 0.1 was used.
We next assessed differences in the abundance of individual microbial species between colonization groups (that is, S. Tm favouring, S. Tm restricting and S. Tm neutral) and untreated controls. We transformed the unrarefied ASV abundances using the centred log-ratio to account for the compositional nature of the sequencing data. Positive centred log-ratio values imply that an ASV is more abundant than average; conversely, negative values imply that the ASV is less abundant than average. Low-biomass samples were removed. Next, we fitted linear regression models to determine which species were differentially abundant between each colonization group and controls using the R package MaAsLin2 (v.1.13.0)66. We included normalized OD578 as a covariate to account for the biomass of the community, with the OD578 values of control samples set to 1. P values were adjusted using the Benjamini–Hochberg method and a significance threshold of 0.1 was used.
Evaluation of metabolic overlap between S. Tm and members of the synthetic community
We estimated potential niche overlap between S. Tm and each member of Com21 by calculating the competition and complementarity indices using PhyloMint (v.0.1.0)67. The metabolic competition index is a proxy of the metabolic overlap of two species; this index is a nonsymmetric measure and it is calculated on the basis of the number of compounds required but not synthesized by both species68 Conversely, the metabolic complementarity index is a proxy for potential syntrophy between species; this index is a nonsymmetric measure calculated based on the number of compounds that one species produces that the second species requires but cannot synthesize68. In brief, PhyloMint takes as input the whole genome sequence of each strain, which it uses to obtain a genome-scale metabolic model with CarveMe (v.1.5.1)69, extracts the metabolite seed sets and calculates the competition and complementarity indices.
Computational processing and analysis of S. Tm and Com20 transcriptomes
We used the nf-core taxprofiler pipeline (v.1.2)70 to pre-process and taxonomically classify the raw reads. In brief, the pipeline used fastp (v.0.23.4)71 for adapter trimming and complexity filtering and Bowtie (v.2.5.2)72 for the removal of eukaryotic contaminant reads, including the human genome (references retrieved from Zenodo: https://doi.org/10.5281/zenodo.4629921). Then, clean reads were taxonomically classified using Kraken2 (v.2.1.3)73 and Bracken (v.2.9)74 against a GTDB-formatted database based on the Unified Human Gut Genome catalogue75 (available at http://ftp.ebi.ac.uk/pub/databases/metagenomics/mgnify_genomes/human-gut/v2.0.2/). We retained the clean reads, which we then used as input for the nf-core metadenovo pipeline (v.1.0.1; available at https://nf-co.re/metatdenovo/), using as the mapping reference the whole genome sequences of all Com20 members or S. Tm (Supplementary Table 14). In brief, metadenovo performed gene calling on the whole genomes used as the reference using prodigal, mapped the reads against the predicted genes using BBmap (v.39.01) from the BBTools suite (available at https://sourceforge.net/projects/bbmap/), obtained counts per each gene present in the assembly using the package Subread (v.2.0.1; https://subread.sourceforge.net/) and annotated the genes using EggNOG mapper (v.2.1.9)76 and KOfamscan (v.1.3.0)77.
For S. Tm, we assessed the distribution of transcript abundances across treatments using the TPM values. We then used DESeq2 (v.1.44.0)78 to identify genes differentially expressed between each treatment and the unperturbed community (DMSO controls). We performed log[fold change] shrinkage to account for genes with low expression and high variability. We considered a gene to be differentially expressed if the absolute value of log2[fold change] was >0.585 and the s value was <0.01. We then used the R package clusterProfiler (v.4.12.6)79 to carry out an overrepresentation analysis of the differentially expressed genes based on KEGG annotations. We used variance-stabilized transformed counts of the 500 most abundant genes to perform a principal component analysis.
For Com20, we assessed the distribution of transcript abundances of each species across treatments using TPM values. Given the large variation in expression levels between species across treatments, we were unable to perform a differential abundance analysis. Therefore, for each bacterium on each treatment, we performed an overrepresentation analysis with clusterProfiler and we examined the pathways represented by the 20% most expressed genes on each species based on KEGG annotations. We also determined the fraction of the top 20% highest expressed genes that were previously identified as stress-related markers in bacteria22. In brief, a previous study22 assessed the gene expression profiles of 32 bacterial pathogens across 11 stress conditions; we retrieved the ‘probability to be differentially expressed (PTDEX)’ score from this publication and used it to classify a gene as a marker of stress if its PTDEX ≥ 0.25 in at least 6 stress conditions.
In vivo colonization assays for S. Tm
Animal experiments were approved by the local authorities in Tübingen (Regierungspräsidium Tübingen, H02/20G and H02/21G). Animals were housed under a 12–12-h light–dark cycle at a temperature of 22 ± 2 °C and a relative humidity of 50–56%. Group sizes were determined using power analysis with G*Power. We used 5–6-week-old mice. Male and female mice were housed in separate cages. These cages were then randomly assigned to the experimental groups, ensuring that each group included mice of both sexes. In the in vivo experiments, blinding was not performed because drug-specific side effects needed to be evaluated. However, pathoscoring was conducted in a blinded manner by two independent assessors.
Defined colonized and humanized mice
Germ-free C57BL/6J mice were bred in-house (Gnotobiotic Mouse Facility, Tübingen) under germ-free conditions in flexible film isolators (Zoonlab) and transferred to the Isocage P system (Tecniplast) to perform experiments. We fed the mice with autoclaved drinking water and γ-irradiated maintenance chow (Altromin) ad libitum. We kept mice in groups of 3–4 animals and tracked their health status every day.
For the Com20 model, we used both female (n = 25) and male (n = 14) mice. For the humanized model, we used three female mice and seven male mice to test the effect of the drug on the host before infection, and ten female mice and five male mice for the S. Tm infection. The gut microbiome of these humanized mice contained a mean of 58 ASVs before treatment.
SPF mice
Male (n = 54) SPF C57BL/6J mice (632C57BL/6J) were purchased from Charles River Laboratories at the age of 35–41 days. After delivery, we kept mice in groups of 3 in individually ventilated cages for a 2-week acclimatization period. Mice were fed with autoclaved drinking water and a maintenance diet for mice (Sniff) ad libitum. We performed the experiments in a laminar flow system (Tecniplast BS60) and scored animals every day. The gut microbiome of these mice contained a mean of 199 ASVs before treatment.
Preparation of bacterial communities and colonization of germ-free mice
We prepared Com20 under anaerobic conditions (2% H2, 12% CO2, the rest N2) in a chamber (Coy Laboratory Products). Consumables, glassware and media were pre-reduced at least 2 days before inoculation of bacteria. We grew each strain as a monoculture overnight at 37 °C in 5 ml of their respective growth medium. The next day, we subcultured bacteria 1:100 in 5 ml fresh medium and incubated them for 16 h at 37 °C, except Eggerthella lenta, which was grown for 2 days. We measured the OD578 and mixed bacteria in equal ratios to a total OD578 of 0.5 (OD578 of 0.025 for each of the 20 strains) in a final volume of 10 ml. After adding 2.5 ml of 50% glycerol (with a few crystals of palladium black (Sigma-Aldrich)), 200 µl aliquots were prepared in 2 ml glass vials (Supelco, Ref. 29056-U) and frozen at −80 °C. We used frozen vials within 3 months.
For the human donor-derived community, we inoculated 500 µl of the stool mixture into 100 ml mGAM and incubated it overnight at 37 °C under anaerobic conditions. The following day, we prepared 200 µl aliquots following the protocol used for Com20 and stored the vials at −80 °C. We used these aliquots within 3 months.
To colonize germ-free mice, cages were transferred to an ISOcage Biosafety Station (IBS; Tecniplast) through a 2% Virkon S disinfectant solution (Lanxess) dipping bath. We kept glycerol stocks of the frozen Com20 community or the complex human microbiome (one per mouse) on dry ice before thawing them during transfer into the IBS. We used the mixtures directly after thawing with a maximal time of exposure to oxygen of 3 min. We colonized mice by oral gavage volume of 50 µl and gavaged them again after 48 h using the same protocol. The IBS was sterilized with 3% perchloracetic acid (Wofasteril, Kesla Hygiene).
To monitor the in vivo stability of Com20 in gnotobiotic mice, we collected fresh faecal samples from every defined colonized mouse after 2, 6, 28 and 57 days after the second colonization. DNA was extracted using a DNeasy PowerSoil HTP 96 kit, and community composition was analysed by 16S rRNA amplicon sequencing, as described above.
In vivo S. Tm challenge
The day before infection, we inoculated a S. Tm culture in LB broth supplemented with 0.3 M NaCl using colonies from a plate and grew the culture for 12 h on a rotator (Stuart, SB3, speed 9) at 37 °C. Fifty microlitres of S. Tm was subcultured in 5 ml LB broth supplemented with 0.3 M NaCl and incubated for 3 h in the same conditions. We washed 1 ml of the subculture twice with 1 ml of ice-cold PBS in a 2 ml Eppendorf tube by centrifugation at 4 °C and 14,000g for 2 min. The pellet was resuspended in 1 ml of ice-cold PBS and kept on ice until oral administration. Mice were infected with a S. Tm load of 5 × 106 CFU in 50 µl PBS.
S. Tm growth inhibition in defined colonized mice
To determine whether Com20 confers colonization resistance in mice, we colonized germ-free mice with Com20 for 28 days. We then treated them with 50 µl of 25% DMSO (solvent control) to test for colonization resistance levels in this mouse model. For comparison, we treated conventional SPF mice with a complex microbiome in the same manner as the mice colonized with Com20. The next day we infected all groups with 50 µl of 5 × 106 CFU of S. Tm. After 16–20 h, mice were euthanized by CO2 and cervical dislocation, dissected and their intestinal contents were collected from the colon. We weighed the faecal samples, diluted them in buffer (2.5 g BSA, 2.5 ml Tergitol and 497.5 ml PBS) and plated the samples on MacConkey agar containing 50 µg ml–1 streptomycin. After incubation overnight at 37 °C, we counted colonies of S. Tm.
Treatment with non-antibiotic drugs and infection with S. Tm
Five non-antibiotic drugs were chosen on the basis of the S. Tm challenge assay: clotrimazole (38 mg kg–1), zafirlukast (20 mg kg–1), chlorpromazine (3 mg kg–1), terfenadine (25 mg kg–1) and clomiphene (60 mg kg–1). We dissolved the drugs in 25% DMSO (DMSO and autoclaved drinking water), aliquoted them and stored them in 2 ml glass vials (Supelco, 29056-U) at −80 °C. For every experiment, we prepared fresh drug solutions. Defined colonized (28 days after colonization), humanized (10 days of colonization) and SPF mice were orally gavaged daily for 6 days with 50 µl non-antibiotic drug or 25% DMSO. We collected fresh faecal samples immediately before the first treatment (day 0) and after 6 days of treatment (day 6), immediately before the infection with S. Tm. Humanized mice were infected for 4 days, during which faecal samples were collected daily after infection.
Fifteen to twenty hours afterwards for defined and SPF mice and 4 days afterwards for humanized mice, we euthanized mice by CO2 and cervical dislocation, dissected them and collected intestinal contents from colon and caecum in pre-weighed 2 ml Eppendorf tubes. After weighing the samples, we added 500 µl buffer (2.5 g BSA, 2.5 ml Tergitol and 497.5 ml PBS) and one sterile steel ball (Agrolager, RB-5/G20W) per tube. We collected half a spleen, mesenteric lymph nodes and half a liver lobe in 2-ml Eppendorf tubes containing 500 µl buffer and one steel ball. All samples were lysed with a TissueLyser II (Qiagen) for 1 min at 25 Hz. We plated intestinal contents and organs on MacConkey plates supplemented with 50 µg ml–1 streptomycin, and in case of humanized mice, remaining intestinal content was kept at −20 °C to measure mouse lipocalin-2. We incubated the plates at 37 °C aerobically overnight and counted colonies the next day to determine the CFU per organ.
ELISA of mouse lipocalin-2
We measured lipocalin-2 in humanized mice every day after infection (days 1–4) as a proxy for intestinal inflammation. We collected faecal samples each day after infection, which we centrifuged at 16,900g for 10 min at 4 °C; we transferred the supernatant to new 1.5 ml Eppendorf tubes (see the section ‘Treatment with non-antibiotic drugs and infection with S. Tm’). We stored the supernatant at −20 °C and thawed it no more than three times before use. For the ELISA (Mouse Lipocalin-2/NGAL, R&D Systems, DY1857-05) we used 96-well Maxisorp NUNC plates (Thermo Scientific, 439454) and performed the assay and the analysis according to the manufacturer’s instructions.
Pathoscoring
Histopathological analysis of caecal tissue was carried out as previously described12. The following features were scored: submucosal oedema (0–3), infiltration of polymorphonuclear neutrophils (0–4), loss of goblet cells (0–3) and epithelial damage (0–3). The individual scores were then summed to provide a final pathology score, categorized as follows: 0–3 (no inflammation), 4–8 (mild inflammation) and 9–13 (severe inflammation). Scoring was conducted by two independent, blinded examiners. Statistical analysis was performed using generalized linear mixed models (lme4 v.1.1-35.5 package), with the animal identifier included as a random effect to account for non-independence owing to score replicates. Maximum likelihood estimation was carried out using the Laplace approximation. A generalized linear model with an appropriate link function was used to accommodate log-normally distributed errors, as determined from model residuals. Model convergence was ensured using iterative derivative-free bound optimization through quadratic approximation (bobyqa). The package emmeans (v.1.10.6) was used to extract estimates of marginal means and contrasts from the model. Fixed-effects coefficients and post hoc contrasts were evaluated using two-sided Wald z-tests.
Immunohistochemistry
We used 25 ml 4% paraformaldehyde solution (ROTI Histofix, Carl Roth, P087.2) to fix the caecal tissue of mice for 24 h at room temperature. The next day, tissue was transferred to a new 50 ml Falcon tube containing 25 ml fresh 4% paraformaldehyde solution and was fixed for another 24 h at room temperature. We placed the tissue in embedding cassettes before it was dehydrated, embedded in paraffin and cut to 2 µm tissue slices. Slices were deparaffinized and rehydrated, antigens were retrieved with Bond citrate solution (AR9961, Leica) and Bond EDTA solution (AR9640, Leica). We incubated the slides with primary antibodies in Bond primary antibody diluent (AR9352, Leica) followed by secondary antibodies (rabbit anti-rat IgG H&L preadsorbed; 1:1,000 (Abcam) or polymer anti-rabbit poly-HRP-IgG (Leica)) and stained using a Bond Polymer Refine Detection kit (DS9800, Leica) (Supplementary Table 13). The slides were scanned using a whole-slide scanner (Aperio AT2, Leica) at ×20 magnification. The following antibodies were used: anti-CD31 (rat, 1:40; Dako, M0823); anti-RelA (rabbit, 1:400; Novus Biological, NB100-2176); HIF1α (rabbit, 1:500; Novus Biological, NB100479); CD4 (rat, 1:1,000; Thermo Fisher, 14-9766-82); CD8 (rabbit, 1:400; Cell Signaling, 98941S); CD11b (rabbit, 1:10,000; Abcam, ab133357); CD11c (rabbit, 1:300; Cell Signaling, 97585); F4/80 (rabbit, 1:400, Cell Signaling, 70076); cleaved caspase-3 (rabbit, 1:300; Cell Signaling, 9661); Ki-67 (rabbit, 1:100; Thermo Fisher, RM-9106-S1); and B220 (rat, 1:3,000; BD, 553084). Slide scans were imported into QuPath (v.0.5.1)80 for quality control and stain vector normalization. The caecal mucosal area (comprising epithelial cells and the lamina propria, delineated by the muscularis mucosae) was segmented to obtain approximately six representative fields of view per animal per staining. This resulted in 664 unique fields of view for the experiment without S. Tm infection and 898 for the infection experiment. On average, we analysed 30,610 cells per field (95% CI: 29,007–32,213) in the infection experiment and 15,759 cells per field (95% CI: 14,821–16,697) in the non-infection experiment, which totalled 22,621,076 analysed cells for the infection experiment and 8,573,164 for the non-infection experiment. For markers in which supracellular signals were of interest, Fiji (v.1.10.6) was used to generate segmentation masks, which were then used to extract morphometric data using the BioVoxxel Toolbox (v.2.6.0) extended particle analyzer81. The primary readout was defined as either the number of positive cells per mm2, determined through histogram-informed thresholding with additional correction for residual staining to minimize bias from sample preparation, or, for markers without a clear biological dichotomy, the quantitative characterization of pixel intensities per cell. These intensities were hierarchically summarized to derive the average cell staining intensity per field of view. To account for potential biases due to variations in tissue cellularity (for example, inflammatory cell infiltration), both readouts (positive cells per mm2 and average cell staining intensity) were also analysed using the number of nuclei instead of surface area as a covariate. Generalized linear mixed models were fitted using lme4 (v.1.1-35.5) to reflect non-independence of observations due to the hierarchical structure of the data82. Maximum likelihood estimation was performed using the Laplace approximation and linear regression was generalized using appropriate link functions for normally or log-normally distributed errors, as determined from model residuals. Model convergence was ensured through iterative derivative-free bound optimization by quadratic approximation (bobyqa) for maximum likelihood estimation. The package emmeans (v.1.10.6) was used to extract estimates of marginal means and contrasts from the model. Fixed-effects coefficients and post hoc contrasts were evaluated using two-sided Wald z-tests.
Assessment of drug treatment on the microbiome composition of gnotobiotic and SPF mice
We assessed the effect of drugs on the composition of the gut microbiome of gnotobiotic, humanized and SPF mice after 6 days of treatment and compared with untreated controls. For this comparison, we carried out analysis of covariance incorporating the abundance of each ASV at day 0 (pretreatment), thus estimating the baseline adjusted difference between groups at day 6. Analyses of covariance models were fitted using a multiple linear regression; robust standard errors were calculated using the R package sandwich (v.3.0-2)83, which were then evaluated by a coefficient test as implemented in the R package lmtest (v.0.9-40)84. To account for the compositional nature of the sequencing data, we transformed ASV abundances with the centred log-ratio. P values were adjusted using the Benjamini–Hochberg method and a significance threshold of 0.1 was used.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.