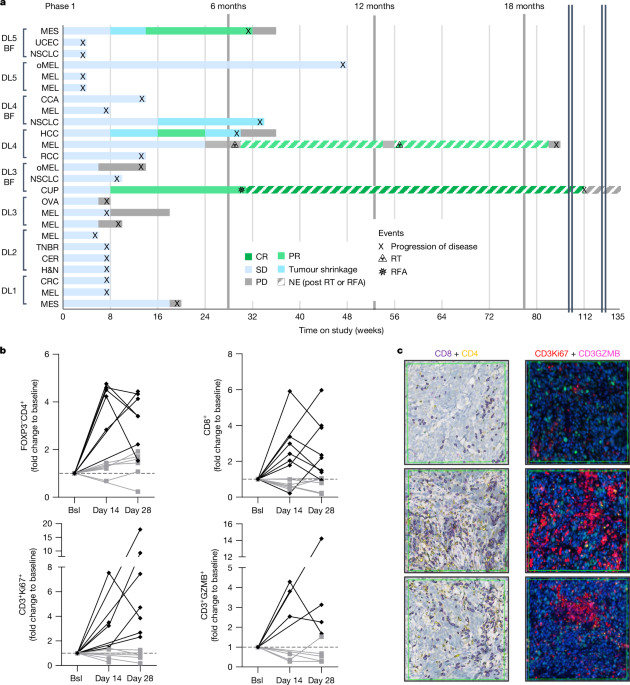
Patients and treatment
The trial was termed GDFATHER, for GDF-15 antibody-mediated human effector cell relocation (ClinicalTrials.gov NCT04725474). This study was conducted in compliance with the International Council for Harmonisation E6 guideline for Good Clinical Practice and the Declaration of Helsinki. Complete and signed written informed consent was obtained from patients for inclusion in the study. The protocol was approved by the regulatory authorities in Germany (Paul-Ehrlich-Institut), Spain (Agencia Española de Medicamentos y Productos Sanitarios) and Switzerland (Swissmedic) and the local ethics committees in charge of the clinical trial site: Comité de Ãtica de la Investigación con Medicamentos, Pamplona, Spain; Swissethics, Cantonal Ethics Committee, Zurich, Switzerland; and the Ethics Committee of the University of Würzburg, Würzburg, Germany. The redacted protocol is available in the Supplementary Information and publicly available at the Clinical Trials Information System, a database set up and maintained by the European Commission.
Patients were eligible for enrolment if they had advanced-stage, relapsed or refractory solid tumours; had exhausted available approved standard treatments, including being relapsed or refractory to prior anti-PD-1 or anti-PD-L1 treatment; (for phase 1; part A and selected phase 2a cohorts) presented with biopsy-accessible tumour for serial biopsy taking; were 18 years or older; and had signed the informed consent form.
The study consisted of two parts, part A (phase 1 (dose escalation)) being a classic â3â+â3â dose-escalation study and part B (phase 2a (expansion)) to explore the antitumoural activity of the combination. In part A of the study, a total of 25 patients were enrolled to receive five predefined DLs and received escalating doses of visugromab intravenous infusion (0.3, 1, 3, 10 and 20âmgâkgâ1) every 2 weeks. The first three patients for each DL received visugromab as monotherapy for one cycle (14âdays) followed by the combination of visugromab and nivolumab. Nivolumab was also administered as an intravenous infusion at 240âmg every 2 weeks. Triple tumour biopsies were taken at baseline, day 14 and day 28.
In part B of the study, up to 5 cohorts with up to 27 participants per cohort with defined tumour entities expected to be GDF-15 dependent were treated with a recommended phase 2 dose, and safety and preliminary efficacy of CTL-002 monotherapy and the combination were evaluated further. To rule out significant visugromab-independent antitumoural activity of nivolumab, patients were included only if they were relapsed or refractory to prior, approved anti-PD-1- or anti-PD-L1-containing treatment as per defined, strict criteria. Patients were enrolled only if they had a minimum of 12 weeks of continuous prior exposure to anti-PD-1 or anti-PD-L1, and their relapse or progression on prior approved anti-PD-1- or anti-PD-L1-containing treatment had occurred while this anti-PD-1 or anti-PD-L1 treatment was ongoing. Based on data in the literature, the expected rate of patients responding to retreatment with an approved PD-1 or PD-L1 agent such as nivolumab in monotherapy in these populations is â¤5% for NSCLC10,11,12,13,14,15.
The study was initiated in December 2020 and the first patient was enrolled on 9 December 2020. As of October 2023, phase 1 of the study has been completed and phase 2a is ongoing with a total of 174 patients enrolled overall in the study.
Endpoints
The main endpoints were safety of visugromab (CTL-002) in combination with nivolumab and antitumoural activity. Safety parameters evaluated for this purpose were the number of participants with adverse events, including serious adverse events; clinical laboratory data; vital signs; electrocardiograms; physical examination (including neurological assessment); and Eastern Cooperative Oncology Group performance status. For phase 1 (part A) dose-limiting toxicities and maximum tolerated dose were also evaluated using National Cancer Institute Common Terminology Criteria for Adverse Events (NCI-CTCAE) Version 5.0. Investigator-assessed evaluation of the antitumour activity was performed according to RECIST V1.1 including the assessment of the proportion of participants with tumour shrinkage (declared if RECIST V1.1-defined reduction in target lesions was â¥5% or more and <30%), a confirmed PR (â¥30% reduction in target lesions) and/or CR, and ORR and various related parameters such as time to response, time to progression and DOR (measured from the time point of signing the informed consent). Secondary and exploratory endpoints included PK, pharmacodynamics (for example, degree of GDF-15 neutralization achieved and change in immune-cell number and composition in the tumour tissue) and cachexia-related parameters such as change in weight.
TCGA data and correlation analysis
The analysis consisted of 30 different cancer types with a varying number of solid tumours analysed. Material included in the TCGA database is derived from primary tumours of untreated (meaning immune checkpoint blockade naive) patients. The full list of abbreviations used, study names and number of samples for solid tumours is available in Extended Data Table 1.
For these indications, gene expression data (RNAseq-v2 raw counts and TPM) were downloaded from http://firebrowse.org on 1 August 2019 (Broad Institute of MIT and Harvard). Duplicated and ambiguous genes were removed, and normalized data were log2-transformed (log2[TPMâ+â1]). Correlation analyses of normalized GDF15 expression with immune-related genes and signatures (Extended Data Table 2) were performed using Spearmanâs rank correlation (test) with the normalized enrichment score from single-sample gene set enrichment analyses or averaged expression levels and visualized as heat maps. GDF15 expression was analysed in different molecular subtypes and in association with clinical parameters in primary tumours of selected indications. Differences in expression between levels of molecular cancer subtypes were tested using a two-sided Wilcoxon rank sum test. Subsequently, Pâvalues were adjusted for multiple testing with the BenjaminiâHochberg method.
Change of estimated immune infiltrates between GDF15 expression groups
Fractions of immune infiltrates and other cell types (including tumour and stromal cells) were estimated through quanTIseq38,39 using the immunodeconv R package40 on RNA-sequencing data (TPM) corrected for purity as determined by ESTIMATE41. Average distribution within a subgroup of patients was computed according to GDF15 expression log2[TPMâ+â1] (low, medium and high terciles) within the respective tumour types. The distribution of immune-cell fractions was averaged over patients in these groups and visualized as a stacked bar plot (including a fraction with other cell types such as tumour and stromal cells).
Software and resources
All calculations, correlations and visualization analyses were performed using the statistical software environment R as well as the resources outlined in Extended Data Table 4.
Measurement of chemokine levels in GDFATHER patient samples
Serum samples for assessment of chemokine levels were taken at screening and each scheduled visit day from cycle 1 until cycle 3, and at the end-of-treatment visit. At dosing days (day 1), the samples were taken within 30âmin before infusion. The serum was isolated using standard procedures and cryopreserved at â80â°C until use. Concentrations of the CXCL9 (MIG) and CXCL10 (IP10) chemokines were determined using validated solid-phase sandwich enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturerâs instructions (R&D Systems; human CXCL9/MIG Quantikine ELISA kit, catalogue no. DCX900; human CXCL10/IP10 Quantikine ELISA kit, catalogue no. DIP100). Both assays were conducted in compliance with the Principles of Good Laboratory Practices regulations.
Measurement of serum visugromab levels in GDFATHER patient samples
Serum samples for PK assessment of total visugromab were taken at screening and every treatment cycle within 30âmin before dosing and just at the end of each infusion. The serum was isolated using standard procedures and cryopreserved at â80â°C until use. Concentrations of visugromab were determined using a validated electrochemiluminescence assay method. The PK assay was conducted in compliance with the Principles of Good Laboratory Practices regulations.
Measurement of serum GDF-15 levels in GDFATHER patient samples
Serum samples for GDF-15 assessment were taken at screening and every treatment cycle within 30âmin before dosing, isolated using standard procedures and cryopreserved at â80â°C until use. Samples from patients at screening were analysed for baseline GDF-15 levels using a validated quantitative solid-phase sandwich ELISA kit according to the manufacturerâs instructions (R&D Systems; human GDF-15 DuoSet ELISA Kit, catalogue no. DY957). Total GDF-15 levels (free GDF-15 plus visugromab-GDF-15 complex) during treatment were determined by a validated ECL method, using a custom visugromab non-competitive anti-GDF-15 nanobody as a capture reagent and, following saturation with visugromab, a custom non-competitive anti-visugromab antibody as a detection reagent. Both GDF-15 assays were conducted in compliance with the Principles of Good Laboratory Practices regulations.
Measurement of serum GDF-15 levels in translational patient samples
Tumour, serum samples and patient data used in the translational part of the study were provided by the University Cancer Center Frankfurt (UCT). The study was approved by the institutional review boards of the UCT and the responsible ethical committees at the Goethe University Frankfurt (project number SUG-2-2022).
For analysis of serum GDF-15, archived samples, which were taken within 1â89 days before surgery for patients with invasive bladder cancer (nâ=â34) or invasive upper urinary tract carcinoma (nâ=â3), as well as before treatment start with systemic therapy and within 1 year beforeâafter biopsy for patients with metastatic urothelial cancer (nâ=â13), were provided. The serum was analysed for GDF-15 levels using a quantitative solid-phase sandwich ELISA kit according to the manufacturerâs instructions (R&D Systems; human GDF-15 DuoSet ELISA Kit, catalogue no. DY957).
Multiplex histological analyses of GDFATHER patient biopsies
Immunohistochemical (IHC) staining and immunofluorescence staining were performed on 4-µm formalin-fixed and paraffin-embedded (FFPE) sections of tumour tissue from consenting patients. Sections were deparaffinized and pretreated by protease digestion or heat-mediated treatment before antibody incubation. Before conducting any IHC or immunofluorescence analysis, a histology assessment was performed by a board-certified medical pathologist on slides stained with haematoxylinâeosin. The slides were stained using the Ventana HE 600 automated staining system (Roche Diagnostics) and scanned using bright-field imaging on the Leica Aperio AT2 platform (Leica Biosystems) using Scanscope software (console, v102.0.7.5; controller, v102.0.8.60) and a UPlanSapo 20Ã/0.75 objective (plus Doppler lens for Ã40 images). The assessment consisted of confirmation of tumour type, assessment of histological features, presence of invasive margin, and determination of percentage of necrotic area on the whole slide and in the malignant lesion area.
Evaluation of intratumoural pro-GDF-15 expression levels in human FFPE samples
Determination of the intratumoural pro-GDF-15 expression levels was carried out applying a rabbit polyclonal anti-GDF-15 antibody (Sigma Aldrich; product no. HPA011191) as the primary antibody for automated staining using the Ventana BenchMark Ultra platform (Roche; software version no. 12.3.1 and 12.5.4). The binding of the anti-GDF-15 antibody was visualized using a secondary horseradish peroxidase (HRP) enzyme-conjugated antibody (Roche Diagnostics; Ventana optiView Universal DAB Detection Kit, catalogue no. 760-700). This specific antibodyâenzyme complex was then visualized with a precipitating enzyme reaction product. Evaluation was carried out by a board-certified pathologist. Evaluation considered cytoplasmic staining (and in cases in which it was applicable, also membranous staining) of tumour cells. Cytoplasmic staining was assessed in four staining intensity categories, ranging from 0 (no staining) to 3+ (intensive staining). The percentage of stained cells per staining intensity category (0 to 3+) was recorded. This classification provided the basis for the calculation of the Hâscore, which describes the GDF-15 protein levels in the tumour. The Hâscore was determined by adding the results of multiplication of the percentage of cells with their respective intensity values as follows:
$${\rm{H}}\,{\rm{score}}=[1\times (\text{percentage}\,\text{of}\,\text{intensity}\,1)]+[2\times (\text{percentage}\,\text{of}\,\text{intensity}\,2)]+[3\times (\text{percentage}\,\text{of}\,\text{intensity}\,3)]$$
IHC evaluation of PD-L1 protein expression in human FFPE samples
Determination of the PD-L1 protein expression level was performed equivalent to the pro-GDF-15 assessment using a rabbit monoclonal anti-PD-L1 (SP263) antibody (Roche Diagnostics; catalogue no. 790-4905) as a primary antibody for automated staining following the manufacturerâs protocol. A rabbit IgG monoclonal antibody (Roche Diagnostics; catalogue no. 790-4795) was used as an isotype control. PD-L1 IHC was evaluated by a board-certified pathologist applying TPS and combined positive score as previously described42.
Multiplex IHC evaluation of CD4, CD8 and FOXP3 expression
CD8, CD4 and FOXP3 IHC staining was performed as a triplex IHC assay on one FFPE tissue slide. CD8 staining was carried out using a monoclonal mouse anti-human CD8 antibody (clone C8/144B; Agilent Technologies; product no. M710301-2) as a primary antibody. CD4 staining was carried out using a monoclonal rabbit anti-human CD4 antibody (clone SP35; CellMarque; product no. 104R), and FOXP3 was stained using a monoclonal rabbit anti-human FOXP3 antibody (clone SP97; Abcam; product no. Ab99963).
Depending on the primary antibody, rabbit- or mouse-specific hydroxyquinoxaline (HQ)-conjugated secondary antibodies (Roche Diagnostics; anti-mouse HQ, product no. 760-4814, anti-rabbit HQ, product no. 760-4815) or rabbit-specific nitropyrazole (NP)-conjugated secondary antibodies (Roche Diagnostics; anti-rabbit NP, product no. 760-4817) were used to allow signal amplification. The binding of the specific primary antibody was visualized using a secondary HRP-conjugated antibody or alkaline phosphatase-conjugated antibody (Roche Diagnostics; anti-HQ HRP, product no. 760-4820; anti-NP alkaline phosphatase, product no. 760-4827).
The enzyme coupled to the secondary antibody catalysed a chromogenic reaction at the binding site of the actual primary antibody resulting in teal-, yellow- and purple-coloured precipitation (Roche Diagnostics; DISCOVERY Teal HRP Kit, product no. 760-247; DISCOVERY Yellow Kit, product no. 760-239; DISCOVERY Purple Kit, product no. 760-229). Triplex IHC staining was carried out on a Ventana Discovery Ultra stainer (Roche; software version 12.5.4). For the identification of tumour epithelium, additional staining for pan-cytokeratin IHC analysis was performed on a separate FFPE tissue slide using a mouse anti-human pan-keratin antibody (clone AE1/AE3/PCK26; Roche Diagnostics; product no. 05267145001). For melanoma samples, an additional SOX10 IHC staining was performed on a separate FFPE tissue slide using a monoclonal rabbit anti-human SOX10 antibody (clone SP267; Roche Diagnostics; product no. 760-4968) to support the tumour-stroma separation process in the digital image analysis. Stained slides were scanned on a Leica Aperio AT2 scanner using Scanscope software (console, v102.0.7.5; controller, v102.0.8.60) and a UPlanSapo 20Ã/0.75 objective (Olympus; plus Doppler lens for Ã40 images).
Evaluation of the triplex IHC staining was carried out by pathologist-assisted digital image analysis of representative areas using Visiopharm software (Visiopharm; version no. 2020.01.1 or higher). Visiopharm software uses undisclosed, linear display lookup tables. Visiopharm analysis output gave a readout on the density (positively stained cells per square millimetre) of cells positive for CD4, CD8, FOXP3 and FOXP3 plus CD4 in four different annotated regions of interest (âtumourâ, âtumour stromaâ, âperitumoural stromaâ and adjacent non-neoplastic ânormal tissueâ).
Multiplex immunofluorescence evaluation of CD3, GZMB, Ki67, panCK and SOX10
For the evaluation of CD3, GZMB, Ki67, panCK and SOX10 expression and a nuclear counterstain in human FFPE patient samples, the semi-quantitative UltiMapper I/O T-act kit (Ultivue; product no. ULT20104 or ULT20110) was used. The slides were stained on a Leica Bond RX (Leica Biosystems; software version Bond 6.0.0.431 or higher), and scanned on the Zeiss Axio Scan Z1 fluorescence scanner (Hamamatsu Orca Flash, v4.0, camera (Hamamatsu Photonics); Colibri7 LED light source (Carl Zeiss Microscopy)) using Zen Blue (v3.1) software and a Plan-Apochromat 20Ã/0.8 M27 (Carl Zeiss Microscopy) objective. The slides were subsequently analysed by pathologist-assisted digital image analysis using Visiopharm software (Visiopharm; version no. 2020.01.1 or higher). Visiopharm software uses undisclosed, linear display lookup tables. The output of the digital analysis gave information on cell densities (positively stained cells per square millimetre) in four different annotated regions of interest (âtumour cellsâ, âtumour stromaâ, âtumour areaâ (combining âtumour cellsâ + âtumour stromaâ), and in adjacent non-neoplastic ânormal tissueâ, in cases in which it was applicable).
Multiplex histological analyses of biopsies from patients with UC under early-line therapy
Slides were stained with haematoxylin and eosin (Sakura Finetek). IHC staining of GDF-15 (HPA011191, polyclonal, 1:100) was performed manually on 4-µm fresh FFPE slides. Semi-quantitative evaluation of IHC results was performed by a pathologist blinded to clinical data using a semi-quantitative approach using the Hâscore.
The multiplex immunofluorescence analysis on whole-slide images was described previously43. Slides were stained with Opal 7âColor Automation Kits (Akoya Biosciences). We stained a tumour microenvironment panel: panCK (C-11, Abcam), CD45 (polyclonal, Abcam), PD-L1 (SP142, Abcam), αSMA (1A4, Sigma), Ki67 (SP6, Abcam) and vimentin (EPR3776, Abcam); and an immune-cell panel: CD3 (D7A6E, Cell Signaling), CD8 (C8/144B, Dako/Agilent), CD4 (EPR6855, Abcam), FOXP3 (236A/E7, Abcam). The dye 4â²,6âdiamidinoâ2âphenylindole (DAPI; SouthernBiotech) was used for staining of nuclei. Corresponding Opal fluorophore antibodies were used for visualization, and images were taken with the PhenoImager HT imaging system (Akoya Biosciences). Representative areas with urothelial cancer were selected and analysed with the inForm software (Akoya Biosciences). Cells were segmented and a machine learning algorithm in the inForm software was trained to identify cell populations.
Gene expression analysis of GDFATHER patient biopsies
Gene expression was measured using the NanoString nCounter PanCancer IO 360 Panel (NanoString Technologies). The PanCancer IO 360 Panel consists of 770 genes, including 20 housekeeping genes. Tissue samples were placed on glass slides as 4-μm-thick FFPE sections and five slides were subjected to RNA extraction using the RNeasy FFPE kit (QIAGEN; catalogue no. 73504) and quality control by NanoDrop (Thermo Scientific). The analysis of gene expression was conducted on the nCounter PanCancer IO 360 Panel and NanoString (NanoString Technologies) platform. A quality check had been performed using NanoStringQCpro v1.14.0 (NanoString Technologies). Raw data normalization using the R package NanoStringNorm resulted in very similar relative log expression distributions compared to normalization using nSolver v4.0 (NanoString Technologies) with standard settings. No batch effect between different runs or cartridges was observed by principal component analyses. Normalized expression data were log2-transformed, and housekeeping genes were filtered. Differential gene expression analyses were performed using the R/Bioconductor package limma (linear models for microarray and RNA-sequencing data) between visugromab treatment (dayâ14) and pretreatment (baseline) by applying a paired moderated t-test. These analyses were performed for two groups of participants, one group of participants with immune-cell influx (IMM) indicated by a more than twofold increase in the numbers of CD8+ and CD4+ T cells, and another group of participants with less than twofold increases in the same T cell subsets (NOIMM). Differential expression of individual genes between IMM and NOIMM was tested using two-sided Wilcoxon rank sum test and visualized as box plots. Volcano plots were generated using the R package EnhancedVolcano. Over-represented Reactome pathways for significantly upregulated genes on visugromab treatment (day 14) versus pretreatment (baseline) in the IMM group were determined using pathway information from ConsensusPathDB (http://cpdb.molgen.mpg.de/CPDB) and a Fisherâs exact test adjusted for the PanCancer IO 360 gene panel in the statistical software environment R v4.3.1 (R Development Core Team). Pâvalues were adjusted for the number of Reactome pathways with at least two matching genes using the BenjaminiâHochberg method.
PKâPD modelling of the distribution, elimination and interaction of visugromab and GDF-15
The PKâPD model was derived from non-human primates and describes the distribution, elimination and interaction of visugromab and GDF-15 in the serum compartment and the peripheral compartment. The distribution between the two compartments was modelled with an inter-compartmental clearance. Visugromab clearance was modelled as a first-order elimination from the serum compartment. Mass action kinetics was used to describe the binding of visugromab to GDF-15, forming a complex that was assumed to be eliminated at the same rate as free visugromab from the serum compartment.
Model parameters for non-human primates were scaled to humans using allometric scaling with a 3âkg body weight for non-human primates and 70âkg body weight for human. The allometric scaling coefficients were 1 for volumes of distribution, 0.75 for clearance, 0.667 for the inter-compartmental clearance, and â0.25 for rates.
The scaled human PKâPD model was coupled to a tumour model to predict free tumour GDF-15 levels for the first-in-human dose selection, reflecting visugromabâs mechanism of action and GDF-15 biology. The tumour model included a tumour microvasculature compartment with a blood flow rate of 0.2âmlâ1âminâ1âgâ1, assuming a tumour size of 36âg. Visugromab was entering from the serum side, whereas free GDF-15 was released from the tumour, resulting in reported serum GDF-15 levels in patients with cancer of 0.5, 2 and 10ângâmlâ1 (low, medium and severe scenarios). The calculation of free GDF-15 used the duration it takes the blood to flow through the tumour.
The parameters of this PKâPD model were estimated using total visugromab and total GDF-15 concentration measurements from the dose-range-finding and Good Laboratory Practice toxicology studies. Once the clinical phase 1 study data were available, the parameters were re-estimated. Both estimations were performed using the SEAM algorithm with Monolix software version 2019R1.
Statistical analysis
This phase 1 and 2a trial was largely evaluated on a descriptive basis as antitumoural activity was unknown for this first-in-human trial.
For the phase 2a cohorts, nâ=â14 response-evaluable participants were to be initially recruited for each tumour indication. Assuming a true response rate of 20%, the probability of observing at least 2 responding patients out of 14 participants was 80%. For an assumed true response of 10%, the probability of observing at least 1/14 responses in the cohort was 77%.
If at least one response was observed, cohort expansion was warranted per the design (5% one-sided α-level, 80% power, 5% maximum response probability of a âpoor drugâ, 20% response probability of a âgood drugâ). An additional nâ=â13 participants were then to be added to a cohort. Observing at least 4/27 responses would confirm that the drug warrants further investigation in that indication.
To assess the statistical significance between two independent groups, a two-tailed MannâWhitney test was performed with a Pâvalue of â¤0.05 deemed as statistically significant. The statistical significance between three or more groups was determined by repeated-measures ANOVA with GeisserâGreenhouse correction followed by Dunnettâs test corrected for multiple comparisons with a Pâvalue of â¤0.05 deemed as statistically significant. The correlation between two parameters was computed using a non-parametric Spearmanâs rank correlation with a 95% confidence interval. All statistical analysis was performed using GraphPad Prism software version no. 10.1.2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.