Fly stocks and husbandry
w1118 (BDSC, 5905; the control strain), Akh-Gal4 (BDSC, 25684), btl-Gal4 (BDSC, 78328), c57-Gal4 (BDSC, 32556), c179-Gal4 (BDSC, 6450), CaLexA (BDSC, 66542), Capa-2A-Gal4 (BDSC, 84597), Capa-2A-LexA (BDSC, 84358), Cg-Gal4 (BDSC, 7011), Debcl-Gal4 (BDSC, 81163), Eip71CD-Gal4 (BDSC, 6871), LexAop-FLP (BDSC, 55819), LexAop-Gal80 (BDSC, 32214), LexAop-myr::GFP (BDSC, 32209), Mef2-Gal4 (BDSC, 27390), Mhc-GFP (BDSC, 38462), nSyb(R57C10)-Gal4 (BDSC, 90854), OK6-Gal4 (BDSC, 64199), R9F03-Gal4 (BDSC, 40734), R16E04-Gal4 (BDSC, 47326), R18D09-Gal4 (BDSC, 48814), R18D09-LexA (BDSC, 54752), R20C04-Gal4 (BDSC, 48882), R22H11-Gal4 (BDSC, 48043), R53E05-Gal4 (BDSC, 48196), R54A09-Gal4 (BDSC, 50460), R82E06-Gal4 (BDSC, 40149), αTub84B-FRT.Gal80.FRT (BDSC, 38879), TubP-Gal4 (BDSC, 5138), TubP-Gal80TS (on the third chromosome) (BDSC, 7017), TubP-Gal80TS (on the second chromosome) (BDSC, 7019), UAS-Ca-α1D RNAi (BDSC, 33413), UAS-Ca-α1T RNAi (BDSC, 39029), UAS-Ca-β RNAi (BDSC, 43292), UAS-cac RNAi (BDSC, 77174), UAS-Capa RNAi (3) (BDSC, 28345), UAS-GCaMP6s (BDSC, 42746), UAS-Kir2.1 (BDSC, 6596), UAS-mCD8::RFP (BDSC, 32219), UAS-myr::GFP (on the second chromosome) (BDSC, 32198), UAS-myr::GFP (on the third chromosome) (BDSC, 32197), UAS-PK1-R RNAi (BDSC, 27539), UAS-TrpA1 (BDSC, 26263) and Vglut (OK371)-Gal4 (BDSC, 26160) flies were obtained from the Bloomington Drosophila Stock Center (BDSC). UAS-Capa RNAi (1) (VDRC, 41124; unless otherwise noted, this line was used in all Capa RNAi experiments), UAS-Capa RNAi (2) (VDRC, 101705), UAS-CapaR RNAi (1) (VDRC, 330441; unless otherwise noted, this line was used in all CapaR RNAi experiments), UAS-CapaR RNAi (2) (VDRC, 105556), UAS-dicer2 (on the second chromosome) (VDRC, 60008) and UAS-dicer2 (on the third chromosome) (VDRC, 60009) flies were obtained from the Vienna Drosophila Resource Center (VDRC). Myo1A-Gal4 (DGRC, 112001) and NP1093-Gal4 (DGRC, 103880) flies were obtained from the Kyoto Stock Center (Department of Drosophila Genomics and Genetic Resources, Kyoto Institute of Technology). CapaR-Gal427, tsh(c724)-Gal451 and Uro-Gal452 flies were obtained from T. Koyama and K. Halberg. elav-Gal4(3A3), phm22-Gal4 and UAS-grim flies were obtained from Na. Yamanaka. nSyb(R57C10)-Gal80 was obtained from James W. Truman (University of Washington). All of the RNAi lines used for the in vivo RNAi screens are listed in Supplementary Table 2–4. CapaR-eGFP flies were generated through CRISPR-mediated mutagenesis as described below.
All fly stocks were maintained on standard fly food containing 5.5 g agar (Daishin, P700), 100 g glucose (Showa Sangyo), 40 g dry yeast (Asahi Beer, HB-P02), 90 g cornmeal (Sunny Maize, Yellow-No.4 M), 3 ml propionic acid (Nacalai Tesque, 29018-55), 3.5 ml 10% butylparaben (in 70% ethanol) (Nacalai Tesque, 06327-15) and 1 l of water. All of the experiments were conducted under non-crowded conditions.
For all of the knockdown experiments except for those in Extended Data Fig. 1f, larvae carried the TubP-Gal80TS transgene, which encodes a temperature-sensitive transcriptional repressor17. Unless otherwise specified, these larvae were raised at 18 °C during the early instar stages (L1 or L2). The larvae were then shifted to 29 °C from 0 h AL3E to induce RNAi from L3. For the genetic manipulation of the VaNs, the larvae were reared at 18 °C during the early larval stages and shifted to 29 °C from 12 h after L2 ecdysis to induce grim expression and from 24 h AL3E to induce expression of Kir2.1 or TrpA1.
Unless otherwise specified, progeny from crosses between w1118 and either Gal4 lines or UAS RNAi lines were used as controls.
Preparation of a Ca2+-free HM
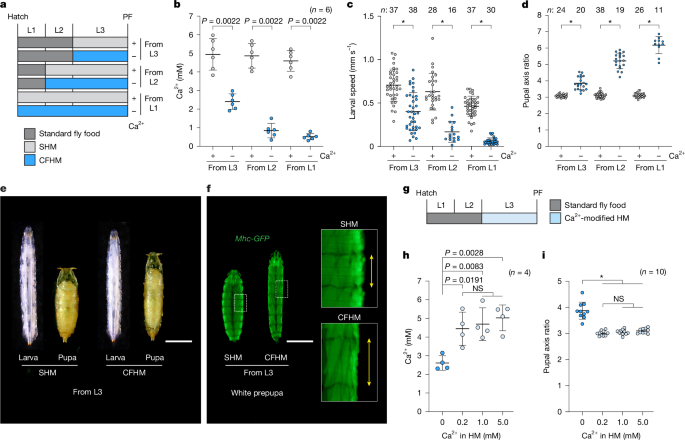
A Ca2+-free diet was prepared by modifying the components of the chemically defined diet for D. melanogaster known as HM8,53,54. Agar, commonly used in the preparation of standard fly food and HM, is rich in minerals, including Ca2+. When we prepared HM using standard agar (Daishin, P700), we detected high levels of Ca2+ even after excluding CaCl2·2H2O from the ingredients (Extended Data Fig. 1a). We therefore measured the Ca2+ concentrations of several purified agar/agarose products (product-A: Nacalai Tesque, 01028-85; product-B: Nacalai Tesque, 01162-15; product-C: Nacalai Tesque, 01163-05; and product-D: Sigma-Aldrich, A9539). We then prepared HM using product-D (Sigma-Aldrich, A9539), which did not contain detectable Ca2+ (Extended Data Fig. 1a). Moreover, we used sodium pantothenate as a substitute for calcium pantothenate. As Ca2+ was undetectable in the resulting HM when we excluded CaCl2·2H2O, we defined it as CFHM. Mg2+-free HM was prepared by excluding MgSO4·7H2O. A complete list of the components used in the preparation of HM is provided in Supplementary Table 1.
Measuring Ca2+ concentrations
For haemolymph samples, wandering L3 larvae were rinsed in Milli-Q water and wiped on Kimwipes. The larval cuticle was carefully torn to release the haemolymph onto a parafilm membrane. Haemolymph (2 µl) was collected from 5 larvae, mixed with 8 µl of ice-cold UltraPure distilled water (Thermo Fisher Scientific, 10977015) and centrifuged at 4 °C for 10 min. The resulting supernatant was stored at −80 °C for later use in the Ca2+ assay.
For whole-body samples, wandering L3 larvae were rinsed in Milli-Q water, wiped onto Kimwipes, transferred individually into tubes, immediately flash-frozen in liquid nitrogen and stored at −80 °C. Dissected tissues or ex vivo cultured tissues were rinsed in UltraPure distilled water and immediately collected in tubes along with UltraPure distilled water. MT samples were collected individually in tubes along with 9 μl of UltraPure distilled water. Other tissues were collected in 45 µl of UltraPure distilled water, flash-frozen in liquid nitrogen and stored at −80 °C until use. Before the calcium assay, whole-body samples were homogenized for 3 min with 50 µl of 0.1 M HCl (diluted with UltraPure distilled water). MT samples were vortexed for 3 min after adding 1 µl of 1.0 M HCl (diluted with UltraPure distilled water to a total of 10 µl). Other tissues were homogenized for 3 min after adding 5 µl of 1.0 M HCl (diluted with UltraPure distilled water to a total of 50 µl). The samples were centrifuged at 4 °C for 10 min, and the resulting supernatants were used for the Ca2+ assay.
Fly food, agar and agarose samples were weighed, placed into sample tubes, melted at 50 °C and used for the Ca2+ assay.
Ca2+ concentrations were measured with the QuantiChrom Calcium Assay kit (Funakoshi, DICA-500) according to the manufacturer’s instructions with the included Ca2+ solution as a standard. Then, 2 μl of each sample and the standard samples were added to a 96-well plate (Thermo Fisher Scientific, 269620) with 100 µl of working solution. After incubation for 3 min at room temperature, the optical density at 612 nm was measured using the Multiskan GO (Thermo Fisher Scientific). For fly food, agarose and agar samples, the assay was performed using prewarmed working solution to prevent solidification. Multiple samples were collected in duplicate and included in the analysis for each experiment.
Analysis of pupal axis ratio
Pupae were affixed to glass slides using double-sided tape, dorsal side up. Images were captured alongside a ruler under a dissecting microscope equipped with a Flexacam C3 12MP microscope camera (Leica). The pupal length and width were analysed using Fiji (ImageJ2, v.2.16.0, National Institute of Health; https://imagej.net/fiji)55. The pupal length was measured from the anterior-most point to the junction of the posterior spiracles. The pupal width was measured along the plane perpendicular to the midpoint of the pupal length axis. Although female pupae were slightly larger than males, we did not consider animal sex because both males and females are known to exhibit equal pupal axis ratios11.
Analysis of muscle contraction activity
Larvae (Mef2-Gal4>UAS-GCaMP6s) were reared on either SHM or CFHM containing red dye (Kyoritsu Foods, 4901325001245) from 0 h AL3E. Prepupal wandering L3 larvae, from which red dye food had been completely purged from the gut, were then used for analysis. Larvae were rinsed with Milli-Q water and dried using Kimwipes. One larva reared on SHM and one larva reared on CFHM were placed side by side on a 35-mm glass-bottom dish (IWAKI, 3910-035, 27 mm glass), and GCaMP imaging was performed simultaneously for 5 min using a Nikon SMZ25 stereomicroscope equipped with a Digital Sight 10 camera (Nikon). At least three independent experiments were performed, with different larvae used for each recording. Only larvae that underwent PF within 1 h after imaging were included in the analysis. In the Ca2+ imaging time-series data, the onset times of body wall peristaltic waves, propagating from posterior to anterior or vice versa, were labelled through visual observation using ImageJ. Propagations across more than three segments were classified as peristaltic waves. The wave frequency was calculated by taking the inverse of the time interval between the onsets of two successive waves. The maximal GCaMP fluorescence signal intensity within the larval body for each wave was measured using Fiji (v.2.16.0) and R (v.4.4.2).
EGTA injections
Larvae (w1118) were reared on standard fly food containing red dye (Kyoritsu Foods, 4901325001245) from 0 h AL3E. Then, prepupal wandering L3 larvae, from which red dye food had been completely purged from the gut, were used for injection. To confirm the injection of the solution into the larval body, 1x phosphate-buffered saline (PBS) (Nacalai Tesque, 27575-31) was coloured blue by adding Erioglaucine disodium salt (final conc. 0.1 g per 10 ml) (Sigma-Aldrich, 861146). Under a dissecting microscope, wandering L3 larvae affixed to glass slides with double-sided tape were injected with EGTA (BMS, BR-401201271) diluted in the blue-coloured PBS. Glass needles were made from glass capillary filament (Narishige, GD-1) using a puller (Narishige, PC-10), and the tips were sharpened using a microgrinder (Narishige, EG-401). Although the exact volume could not be controlled due to injector limitations, each larva was injected with a volume of approximately 100–200 nl. After the injected larvae were rinsed with water to remove any residual glue, they were transferred to vials containing standard fly food and those that successfully formed pupae were analysed to determine their pupal axis ratios.
Larval locomotion assay
Wandering L3 larvae were rinsed in Milli-Q water and wiped on Kimwipes to dry them. Subsequently, 10 larvae were placed together in the centre of a 1% agar plate with a 9-cm diameter (Sansei Medical, 01-013). Water (10 μl) was then added to the larval aggregate in the centre of the plate. The entire plate was recorded for 3 min from above using a fixed-position video camera (SONY, FDR-AX45) with an in-frame ruler. At least three independent experiments were conducted for each genotype, with ten larvae recorded simultaneously in each experiment. Different larvae were used for each recording, resulting in a minimum of 30 larvae recorded for each genotype. The resulting video files were edited using iMovie (v.10.3.5; Apple), Adobe Photoshop (v.24.7.4) and Adobe Premiere Rush (v.2.10.0). The coordinates of the centres of individual larvae in the binarized videos were recorded. Except for the larvae reared on the Ca2+-free diet from the first instar, individual larvae were tracked using the FIMTrack software56. For the larvae reared on the Ca2+-free diet from the first instar, the centres of the larvae were labelled manually using Fiji (ImageJ2, v.2.16.0). This was required because the larvae had such low locomotor activity that they did not separate well enough for FIMTrack to detect individual larvae. Larval locomotor speed was obtained by dividing the distance each larva moved in 10 s by 10 s. The speed of individual larvae was calculated using Python 3.9 by averaging 2–4 speed measurements along a consecutive trajectory.
Analysis of the larval survival rate
The larval survival rate was calculated as the proportion of larvae developed into the pupal stage in each vial. Larvae were transferred into small food vials containing HM at a density of 25 larvae per vial for newly moulted L2 and L3, or 50 larvae per vial for newly hatched L1. The vials were maintained in a humidified incubator, and the number of individuals that successfully formed pupae in each vial was recorded.
Total RNA extraction and RT–qPCR analysis
Animals or dissected tissues from white prepupae (w1118) were collected in 1.5 ml tubes and immediately flash-frozen in liquid nitrogen. Total RNA from animals or tissues was extracted using TRIzol (Thermo Fisher Scientific, 15596-026) according to the manufacturer’s instructions. cDNA was generated from purified total RNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, FSQ-301) according to the manufacturer’s instructions. RT–qPCR was performed on the CFX Duet Real-Time PCR machine (Bio-Rad) using THUNDERBIRD Next SYBR qPCR Mix (TOYOBO, QPX-201). For absolute mRNA quantification, serial dilutions of pGEM-T (Promega, A362A) plasmids containing the coding sequences of a target gene or of rp49 were used for standards. After the molar amounts were calculated, the transcript levels of each target mRNA were normalized to the rp49 levels in the same samples. Three separate samples were collected for each experiment and duplicate measurements were conducted. The following primers were used: Capa forward, 5’-AGAAGAACCGTGACCGTTCCGAGG-3’; Capa reverse, 5’-TCGTGGTCCGTCTCAGCTGTACTG-3’; CapaR forward, 5’-TTCGTGTGCTGGTTCCCGTTCCAC-3’; CapaR reverse, 5’-TCCCGCAATCGAGAAGAGTGCCTC-3’. The primers to detect rp49 levels were previously reported57.
Immunostaining
Tissues were dissected in 1× PBS (Nacalai Tesque, 27575-31), fixed with 4% paraformaldehyde (PFA) (Nacalai Tesque, 02890-45) in PBS (4% PFA/PBS) containing 0.1% Triton X-100 for 20 min at room temperature and then washed multiple times with PBS containing 0.1% Triton X-100 (PBST). Tissues were blocked with 5% normal goat serum (NGS) (Sigma-Aldrich, G9023) in PBST for at least 1 h at room temperature, incubated overnight at 4 °C with primary antibodies mixed in PBST containing 5% NGS, washed multiple times with PBST, incubated for 2 h at room temperature with secondary antibodies mixed in PBST containing 5% NGS and again washed multiple times with PBST. When necessary, DNA was stained with DAPI (Sigma-Aldrich, D9542) at 1:2,000 and F-actin was stained with Alexa Fluor 488 or 568 Phalloidin (Thermo Fisher Scientific, A12379 or A12380) at 1:200 for 30 min at room temperature. After washing, tissues were mounted in Vectashield H-1000 (Vector Laboratories) and observed using the Zeiss Axio Imager M2 equipped with ApoTome.2 or the Zeiss LSM 700 confocal microscope. The specificity of the signals was established by comparison with appropriate controls.
Chicken anti-GFP (1:2,000 dilution, Abcam, ab13970) and rabbit anti-RFP (1:2,000, Medical and Biological Laboratories, PM005) were used as primary antibodies. Rabbit anti-Capa precursor antibodies (1:2,000) and guinea pig anti-Capa mature peptide antibodies (1:500) were generated by BioGate. The rabbit anti-Capa precursor antibodies were generated against the peptide NH2-LDGIYGDASQEDYNEADFQ-COOH. The guinea pig anti-Capa mature peptide antibodies were generated against the peptide NH2-GLYAFPRV-CONH2, which is identical to Capa-1 and matches all of Capa-2 except for one amino acid (Y), suggesting that the resulting antibodies can recognize both Capa-1 and Capa-2. Alexa Fluor 488 goat anti-chicken (1:1,000, Thermo Fisher Scientific, A32931), Alexa Fluor 555 goat anti-rabbit (1:1,000, Thermo Fisher Scientific, A32732) and Alexa Fluor 555 goat anti-guinea pig (1:1,000, Thermo Fisher Scientific, A21435) were used as secondary antibodies.
Generation of a C-terminal eGFP-tagged CapaR CRISPR knock-in line
An eGFP-tagged CapaR line (CapaR-eGFP) was generated by WellGenetics using a slightly modified published method58. In brief, gRNA sequences (AAGGTTGTTCTCGACGTCCT[TGG]) were cloned into U6 promoter plasmids. Two homology arms and an eGFP-3xP3-RFP cassette containing eGFP and a floxed 3xP3-RFP were cloned into pUC57-Kan as a repair donor template. CapaR-targeting gRNAs and hs-Cas9 were supplied in DNA plasmids, together with the donor plasmid, for microinjection into embryos of a w1118 control strain (LWG228). F1 flies carrying the selection marker indicating the presence of 3xP3-RFP were further validated by genomic PCR and sequencing. CRISPR generated a break in CapaR that was then repaired by replacement with the eGFP-3xP3-RFP cassette. The resulting eGFP-tagged CapaR line was homozygous viable and did not exhibit an elongated pupal phenotype, suggesting that the tagged CapaR remained functional.
SEM analysis of MT granules and analysing their ion composition using EPMA
A-MTs were dissected in 1× PBS, rinsed with UltraPure water and immediately collected on a carbon-coated film along with UltraPure water. The initial segment of the A-MT was carefully torn to release the granules in its lumen. Then, after discarding the A-MT tissue, the granules were immediately dried on a carbon-coated film. Before analysis, the granules on carbon-coated film attached to a copper grid were vacuum processed and coated with carbon. The granules were analysed using SEM and ion-composition analysis using a field emission electron probe microanalyzer (FE-EPMA) (JEOL, JXA-8530F) operated at a voltage of 10.0 kV. This analysis revealed the ion composition of the granules, excluding their oxygen and carbon content. Each measurement was performed at least three times on different granules. For the oxygen and carbon composition analysis, samples were collected on copper-coated films without adding a carbon coating. The measured Ca2+ values were used as standard values, and the measured oxygen and carbon values were included in a recalculation of the composition of all ions.
Peptide synthesis and ex vivo culture of MTs
Mature amidated Capa peptides in D. melanogaster—Capa-1 (NH2-GANMGLYAFPRV-CONH2), Capa-2 (NH2-ASGLVAFPRV-CONH2) and PK-1 (NH2-TGPSASSGLWFGPRL-CONH2)—were synthesized by Eurofins Genomics.
Control and CapaR RNAi larvae were reared at 18 °C and shifted to 29 °C from 0 h AL3E to induce RNAi from L3. After 24 h of RNAi induction (24 h AL3E), the larvae were washed with 1× PBS. Then, the two A-MTs and the gut were carefully dissected in Schneider’s Drosophila medium (SDM) (Thermo Fisher Scientific, 21720024) supplemented with 10% FBS (Thermo Fisher Scientific, 10270106). Fat bodies attached to the MTs were carefully removed, and the dissected MT–gut complexes were pre-incubated in SDM containing 10% FBS in a Petri dish (AS ONE, 1-8549-01) for 10 min. Only intact MTs were selected for analysis to prevent leakage of Ca2+ granules from MTs ruptured during the dissection process. After pre-incubation, the MTs were severed at the base where they connect to the gut. Both MTs obtained from the same larva were transferred to separate wells of a 24-well clear flat-bottom tissue culture plate (TPP, 92424). To avoid damaging the MTs during transfer, each MT was transferred along with medium. Finally, six MTs per well were placed in a total of 450 µl of medium. Synthetic Capa peptides (Capa-1, Capa-2 and PK-1) in SDM with 10% FBS were added to a final concentration of 10 μM (50 μl per peptide; total volume, 500 μl per well) and incubated at 25 °C for 30 min. After the incubation, the MTs were washed twice with UltraPure distilled water (Thermo Fisher Scientific, 10977015) and collected individually in tubes along with 9 μl of UltraPure distilled water before being frozen at −80 °C.
Statistics and reproducibility
All statistical analyses were carried out using GraphPad Prism (v.9.5.1). Pairwise comparisons were made using the two-tailed Mann–Whitney U-tests, and multiple samples were compared using the one-way ANOVA with Dunnett’s or Tukey’s multiple-comparison test. In some cases, the two-way ANOVA with Tukey’s multiple-comparison test was performed to examine interactions between variables. All of the experiments were performed independently at least twice using independently reared populations. In each experiment, we analysed three or more specimens. Animals and samples were randomly assigned to experimental groups. Representative images were chosen from at least three options. Data collection and analysis were not performed blinded to the experimental conditions. Although no statistical method was used to predetermine sample size, sample sizes were determined based on significance obtained from previous studies with similar experimental setups. Comparable sample sizes were used in each experiment. No datapoints or animals were excluded from analyses.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.