Ethics
Animal care and surgical procedures were performed according to the Directive 2010/63/EU of the European Parliament, which had been approved by the Ministry of Agriculture, France. The project was approved by the French National Ethics Committee for Animal Experimentation (CEEA) under the number APAFIS 36241-2022032518526906 v4 (for PCP procedure), APAFIS 4111-2016021613253432 v5 (for tritiated nanobodies) and APAFIS 31981-2021060423426200 v3 (for nanobody pharmacokinetics in blood). Behavioural pharmacology in GluN1-KD mice was approved by the Faculty of Medicine and Pharmacy Animal Care Committee at the University of Toronto in accordance with Canadian Council on Animal Care (CCAC) guidelines. All experiments were performed in accordance with relevant named guidelines and regulations.
Animals
C57BL/6J WT mice (21.5 ± 0.3 g) were purchased from Janvier Labs. GluN1-KD mice (Grin1 KD) were generated as previously described26. Congenic C57BL/6J Grin1+/− and 129 × 1/SvJGrin1+/− mice were intercrossed to produce experimental mice (Grin1+/+ (WT) and Grin1-KD) mice as recommended and based on other studies51,52 to minimize the confound of homozygous mutations on each parental strain. Animals were housed under a 12 h–12 h light–dark cycle, at 22 ± 1 °C and 55 ± 10% of relative humidity. All animals had access to water and food ad libitum.
Reagents, cell lines, antibodies and plasmids
HEK293 cells (ATCC, CRL-1573) were cultivated in DMEM or DMEM-GlutaMAX (Life Technologies) complemented with 10% (v/v) fetal bovine serum (Sigma-Aldrich). Absence of mycoplasma was routinely checked using MycoAlert Mycoplasma detection kit (LT07-318, Lonza) according to the manufacturer’s protocol. The pRK5 plasmid encoding WT rat mGlu2 receptor subunit, with an HA-tag and a SNAP inserted just after the signal peptide, was previously described53. All drugs (LY379268 and LY341495) were from Tocris (Bio-techne). HTRF IP-One Gq Detection Kit (62IPAPEB), HTRF anti-6×His monoclonal antibody d2-conjugate (61HISDLA), Tag-lite SNAP-Lumi4-Tb Labelling Reagent (SSNPTBD), HTRF d2-NHS (65D2SABA) were gifted by Revvity. Monoclonal rabbit-anti-6×His, 3-3′-diaminobenzidine chromogen solution (DAB) peroxidase substrate kit and ABC peroxidase kit (anti-rabbit-HRP) were purchased from Invitrogen and Vector Laboratories, respectively.
Plasmid of nanobodies constructs
Bivalent nanobodies were obtained by fusing one copy of DN1, DN10 or DN13 to either the N or C terminus of DN13 using one of the two linkers. A 6×His-tag was inserted at the C terminus of the second nanobody for purification purposes. The sequences of the linkers are as follows: hinge HcAb (EPKIPQPQPKPQPQPQPQPQPKPQPKPEP) or (GGGGS)3: GGGGSGGGGSGGGGS. The bivalent DN13–DN13 constructs with the two different linkers were synthesized by Genecust and cloned between NdeI and XhoI in a modified pHEN1 vector in which the sequence coding for the G3P protein has been removed. The bivalent biparatopic constructs were generated by PCR and subcloned through silent restriction sites inserted into the framework regions FR1 and FR4. To generate the cDNA of DN13–Fc, the monomeric DN13 sequence was inserted into the pcDNA3.1(+) vector with the secretory signal peptide from human interleukin (IL-2) fused to its N terminus and the sequence encoding the Fc region of human IgG1 at its C terminus.
Production and purification
For large scale nanobody production, plasmids encoding nanobodies were transformed in E. coli BL21DE3 strain (Life technologies). A single colony was grown in 10 ml of LB, supplemented with 100 μg ml−1 ampicillin, 1% (w/v) glucose, overnight at 37 °C with shaking. Then 1 l of LB supplemented with around 10 ml of the preculture was incubated until an optical density at 600 nm (OD600) of 0.6–0.7. The nanobody expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside and bacteria were grown overnight at 28 °C with shaking. Bacteria were collected by centrifugation for 10 min at 5,000g, resuspended in 10 ml per 1 l culture of ice-cold TES buffer (0.2 M Tris, 0.5 mM EDTA, 0.5 M sucrose, pH 8) and incubated for at least 1 h at 4 °C on shaking platform. Then, 20 ml per l of culture of TES/4 (TES buffer diluted four times in water) were added to the solution and further incubated for at least 45 min at 4 °C on a shaking platform. The periplasmic extract was recovered by collecting the supernatant after centrifugation of the suspension for 30 min at 10,000g at 4 °C.
To produce DN13–Fc, HEK293F cells (Expi293 Expression System Kit, A14635, Thermo Fisher Scientific) were cultured at a density of 0.6 × 106 per ml with 180 ml fresh medium in a 2 l culture bottle at 37 °C and 5% CO2, shaken at 200 rpm, as previously described for DN42–Fc54. Of the cells, 1–1.5 × 106 cells per ml were transfected with DN13–Fc plasmid (225 μg in 12 ml OMEM) and PEI (675 μg in 733 ml OMEM). Cells were cultured for 4–7 days at 37 °C and 5% CO2 with 200 rpm shaking. The supernatant was collected after a 10 min centrifugation at 2,000g and 4 °C.
The His-tagged nanobodies from bacteria and HEK293F cells were purified from the periplasmic extract or cell supernatants using Ni-NTA purification (Qiagen) according to the manufacturer’s instructions. Desalting was performed using disposable PD-10 desalting columns (Cytiva) according to the manufacturer’s instructions to obtain the nanobodies in phosphate buffer solution. Endotoxins were removed using Proteus NoEndo S (Generon) according to the manufacturer’s instructions before administration of the nanobodies to animals. Part of DN13–DN1 produced followed a second step of purification by size-exclusion chromatography using gel filtration (ÄKTA, Cytiva) before administration of the nanobodies to animals to reduce further endotoxin content.
LPS quantification
E. coli LPS contamination was assessed by targeted quantification of 3-hydroxy myristic acid (3-OHC14:0) as previously described55 with modifications56. In brief, protein aliquots (100 μl, 200–500 μg) were spiked with 20 pmol of 3-hydroxytridecanoic acid (3OH-C13:0, 1 pmol μl−1 in ethanol) used as an internal standard (IS). The samples were then hydrolysed with 8 M hydrochloric acid (300 μl) for 3 h at 90 °C. Released fatty acids were extracted with 600 μl of endotoxin-free water and ethyl acetate:hexane 3:2 (v/v; 5 ml). The organic phases were dried in vacuo. Dried extracts were finally solubilized in 50 μl ethanol and 3 μl were injected into a liquid chromatography–tandem mass spectrometry system as described previously56. Calibration standards (100 μl PBS) containing 0, 0.5, 1, 2, 4, 8, 16, 32 and 64 pmol 3OH-C14:0 were treated as samples. The area under the curves for 3OHC14:0 and 3OH-C13:0 (IS) was determined, and the area ratios 3OH-C14:0/IS were calculated. A linear calibration was used for the calculations. LPS concentration was calculated assuming a molecular mass of 12,000 g mol−1 and that each LPS molecule contains four molecules of 3OH-C14:0. One endotoxin unit (EU) is considered to be equivalent to 0.1 ng of LPS.
Binding experiments based on TR-FRET
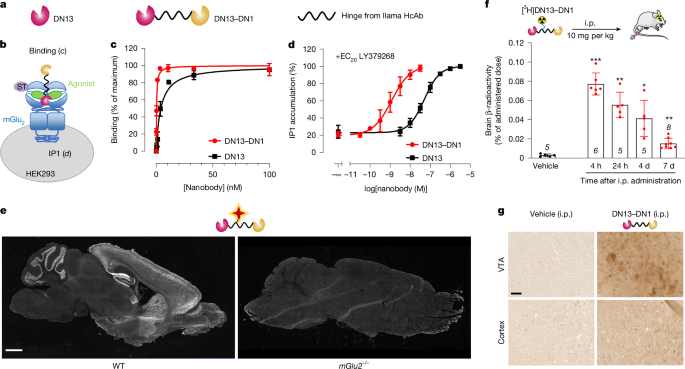
HEK293 cells were transfected with cDNA encoding rat, human or mouse SNAP-tagged mGlu2 receptors and the high-affinity glutamate transporter EAAC1. Then, 24 h after transfection, cells were labelled in 150 mm cell culture plates with 100 nM Tag-lite SNAP-Lumi4-Tb Labelling Reagent in DMEM-GlutaMAX for 1 h at 37 °C, and washed three times with Krebs buffer (10 mM HEPES pH 7.4, 146 mM NaCl, 4.2 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2, 5.6 mM glucose and 0.1% BSA). Cells were detached using cell-dissociation buffer (Sigma-Aldrich) transferred at 10,000 cells per well in a white 384-well plate (Greiner bio-one, PS, F-bottom, small volume). Increasing concentrations of 6×His-tagged nanobodies, together with either 1 μM LY379268 (agonist) or 1 μM LY341495 (antagonist) and with 200 nM of HTRF anti-6×His monoclonal antibody d2-conjugate, were applied to Lumi4Tb-labelled cells in a final volume of 20 μl overnight at 20 °C.
To determine the selectivity of the DN13–DN1 nanobody for the mGlu1–8 receptors, HEK293 cells were transfected with cDNA encoding for human SNAP-tagged mGlu receptors and transferred at 105 cells per well in a black 96-well plate (Greiner bio-one, PS, F-bottom), as previously described54. Then, 24 h after transfection, cells were labelled with 100 nM Tag-lite SNAP-Lumi4-Tb Labelling Reagent in DMEM-GlutaMAX for 1 h at 37 °C, and washed three times with Krebs buffer. Next, 200 nM of 6×His-tagged DN13–DN1 and 300 nM of anti-6×His-d2, and the agonists (1 μM quisqualic acid for mGlu1 and mGlu5; 1 μM LY379268 for mGlu2 and mGlu3; 100 μM L-AP4 for mGlu4, mGlu6, mGlu7 and mGlu8) or the antagonist (10 μM LY341495) were added to the cells in a total volume of 60 μl per well, and incubated overnight at 20 °C.
FRET signals were determined by measuring the sensitized acceptor emission (665 nm) and Tb donor emission (620 nm) using a 50 μs delay and 450 μs integration time after excitation at 337 nm on a PHERAstar FS plate-reader (BMG LabTech) using PHERAstar control software v.5.41. TR-FRET ratio (665 nm/620 nm × 104, patent Cisbio Bioassays, US5,527,684) was calculated for preventing interferences due to medium variability and chemical compound or to normalize experiments when using cells expressing different receptor levels.
Measurements of inositol phosphate
HEK293 cells were co-transfected by electroporation with plasmids encoding rat, human or mouse SNAP-tagged mGlu2 receptors, EAAC1 and GqTop to allow efficient coupling of mGlu2 to the phospholipase C pathway. In total, 100,000 cells were used per well in a black 96-well plate (Greiner bio-one, PS, F-bottom). Then, 24 h after transfection at 37 °C, cells were washed and stimulated with EC20 LY379268 (for human, 0.48 nM; mouse, 0.63 nM; rat, 0.6 nM) or EC20 glutamate (for human, 3.58 μM; mouse, 10.18 μM; rat, 5.42 μM) and various concentrations of nanobodies and incubated for 30 min at 37 °C and 5% CO2. The measurement of IP1 accumulation was determined using the HTRF IP-One Gq Detection Kit (Revvity) according to the manufacturer’s recommendations.
Fluorescent nanobody labelling
Nanobodies were dialysed overnight at 4 °C in carbonate buffer (0.1 mM, pH 9) and incubated (250 μg nanobodies at 2 mg ml−1) with the fluorophore HTRF d2-NHS (Revvity) in phosphate buffer (100 mM pH 7) at a ratio of 6 (fluorophore/nanobody), for 45 min at 25 °C; or in phosphate buffer (50 mM pH 8) and incubated with Lumi4-Tb-NHS (Revvity) in phosphate buffer (100 mM pH 7) at a ratio of 12 for 30 min at 25 °C. Nanobodies were purified by gel filtration column (NAP-5, GE Healthcare) in phosphate buffer 100 mM pH 7. The final molar ratio of fluorophores per nanobody was calculated as the fluorophore concentration/conjugated nanobody concentration, and the conditions were set up for a ratio of between 2 and 3. The concentration of fluorophores in the labelled fraction was calculated for each fluorophore as the OD/ε (OD650 and ε = 225,000 M−1 cm−1 for d2, and OD340 and ε = 26,000 M−1cm−1 for Lumi4-Tb), while that of nanobodies was determined by the OD280. The conjugated concentration was calculated as OD280 − (ODfluo/Rzmax)/ε nanobody with Rzmax = ODfluo/OD280. The average number of fluorophores is 2.5 per DN13–DN1. The purified labelled fraction was supplemented with 0.1% BSA and kept at −20 °C.
Histological processing of brains
WT C57BL/6J mice were purchased from Janvier Labs. Brains from mGlu2−/− mice were provided by J. Gonzalez-Maeso (Virginia Commonwealth University). Mice were anaesthetized (Euthasol Vet 8%, Alcyon) and euthanized by a cardiac perfusion of 4% paraformaldehyde (PFA). Brains were extracted and fixed in a 4% PFA solution (Euromedex) overnight at 4 °C. The brains were rinsed in PBS and cryoprotected in a 30% sucrose solution for 4 days at 4 °C. The brains were then added to OCT (Tissue-Tek, Sakura Finetek), quickly frozen in acetone chilled on dry ice and conserved at −80 °C until use. The frozen brains in OCT were moved to −20 °C 24 h before cryo-sectioning. They were mounted onto a cryostat (Leica) and 16 μm sagittal sections were performed and directly mounted on Superfrost Plus glass slides (Sigma-Aldrich) and kept at −20 °C until use. For immunofluorescence experiments, sagittal brain tissue sections of WT and mGlu2−/− mice were blocked 1 h at 25 °C with a blocking solution (3% BSA, 0.1% Triton X-100 in PBS) and incubated with DN13–DN1–d2 (200 nM), overnight at 4 °C, to label mGlu2 receptors. The sections were washed with PBS, then with distilled water and mounted using Fluoroshield containing DAPI to stain nuclei (Sigma-Aldrich). Images were taken using a slide scanner Axio scan Z1 microscope (Zeiss, Jena) by performing full-section mosaic with a 20× enlargement with Zeiss Zen software 2.3 light. For immunohistochemistry experiments, C57BL/6J female WT mice were injected i.p. either with 10 mg per kg of DN13–DN1 or with an equivalent volume of PBS (150 μl) as a negative control. Then, 24 h after injection, mice were anaesthetized (Euthasol Vet 8%) and euthanized as described above. Ten glass slides per animal, each containing five brain tissue sections, with 80 μm spacing between each section, were rinsed with Tris-buffered saline (TBS) and incubated 1 h at 25 °C with a blocking solution (5% normal goat serum, 0.1% Triton X-100 in TBS). The sections were then incubated with a monoclonal rabbit anti-6×His (1:500, Life Technologies) 2 h at 25 °C and washed in TBS. After blocking endogenous peroxidase (3% H2O2 solution, 20 min at 25 °C), the sections were incubated with a goat anti-rabbit biotinylated antibody (1:100, ABC peroxidase kit, Vector Laboratories). Labelling was revealed using the DAB peroxidase substrate kit (Vector Laboratories). The sections were then washed in distilled water and mounted using Fluoroshield with DAPI. Images were taken using a slide scanner Axio scan Z1 microscope by performing full-section mosaics with ×20 enlargement.
Nanobody quantification in the blood
A quantitative test assessing nanobody concentration has been developed based on the TR-FRET-binding assay on cells expressing SNAP-tagged mGlu2 receptor. To determine the linearity of the assay, increasing concentrations of DN13 or DN13–DN1 were added to 10,000 cells expressing Lumi4Tb-labelled SNAP-tagged mGlu2, 200 nM of anti-6×His-d2 antibody and 1 μM LY379268 in a 384-well plate. After 3 h incubation at 20 °C, the HTRF signal was recorded on a Pherastar FS plate reader and the linearity determined using GraphPad Prism. C57BL/6J WT mice (Janvier Labs) were treated by i.p. administration of 10 mg per kg of DN13, DN13–DN1 or an equivalent volume of PBS (150 μl) and blood samples were collected at different timepoints (1 h, 2 h, 4 h, 6 h, 8 h, 10 h, 12 h and 24 h). The plasma was extracted using a microvette CB 300 K2E (Sarstedt) and frozen until use. Plasma (3 μl) was applied to cells expressing mGlu2 receptor, labelled with Tag-lite SNAP-Lumi4-Tb Labelling Reagent in the presence of 200 nM of HTRF anti-6×His monoclonal antibody d2 conjugate. The FRET signal was determined as indicated for the saturation binding assay.
Nanobody radiolabelling
DN13–DN1 was radiolabelled using bacterial transglutaminase and a synthetic peptide tag, CBz-Gln-Gly-Lys-OH synthetized using manual solid-phase peptide synthesis on a microwave synthesizer (CEM) according to a standard Fmoc-based synthesis protocol. Starting from the Fmoc-Lys(Boc)-Wang resin loaded at 0.32 mmol g−1 (Novabiochem), the synthesis was performed on a 0.015 mmol per reactor scale. Protected amino acids Fmoc-Gly-OH and CBz-Gln-OH (5 eq.) were coupled using HATU (4.5 eq.) and iPr2EtN (10 eq.) in DMF for 20 min at 60 °C. The Fmoc group was deprotected by three successive treatments with 20% piperidine in DMF for 3 min. The peptide was then deprotected and cleaved from the resin by a treatment with trifluoroacetic acid/H2O/triisopropylsilane (95/2.5/2.5) for 2 h at 25 °C. The peptide was then precipitated by dilution into an ice-cold diethyl ether solution, recovered by centrifugation and washed twice with diethyl ether. CBz-Gln-Gly-Lys-OH was tritiated using the following procedure. To a solution of the CBz-protected peptide (87 nmol, 1 eq.) in DMF (50 μl) at 25 °C was added iPr2EtN (870 nmol, 10 eq.) and [3H]N-(propionyloxy)succinimide (87 nmol, 1 eq.). The mixture was stirred at 25 °C for 2 h. The labelled product was purified on a high-performance liquid chromatography system equipped with a Luna Omega C18 (150 × 4.6 mm; 3 μm) column (Phenomenex) with a gradient of 0–100% buffer B in buffer A over 20 min with a flow rate of 1 ml min−1 (buffer A: 0.1% formic acid in H2O; buffer B: 0.1% formic acid in acetonitrile). The radiolabelling of DN13–DN1 through lysine conjugation was carried out in PBS buffer at pH 7.4 using 1 U per 20 nmol of bacterial transglutaminase (Zedira), 2 eq. of the radiolabelled peptide tag and 80 μM of DN13–DN1. The mixture was then incubated for 2 h at 25 °C. After incubation, the labelled protein was purified on Protino Ni-NTA resin (Macherey-Nagel) and then the buffer was exchanged to 0.9% (w/v) NaCl using an Amicon centrifugal filter (Merck) for in vivo experiments. Part of the labelled DN13–DN1 was digested by trypsin to identify Lys121 of the ERK motif in the hinge HcAb, as the conjugation site by MALDI-TOF (Thermo Fisher Scientific) analysis.
In vivo cerebral quantification and localization of the tritiated nanobody and autoradiography
Experiments were conducted on adult female WT C57BL/6J mice (18.8 ± 0.2 g) purchased from Janvier Labs. Mice were injected i.p. with 10 mg per kg of [3H]DN13–DN1 (3 μCi) or vehicle in 50 μl. The mice were euthanized 4 h, 24 h, 4 days or 7 days after administration (only at 24 h for vehicle group), perfused with 0.9% (w/v) NaCl (10 ml per mouse, 200 ml h−1) for brain collection and freezing. Brains were used for either tissue quantification or cerebral localization of β-radioactivity. Quantification of β-radioactivity contained in total brain homogenate was performed using a liquid scintillation analyzer (Tri-Carb 2100TR, Packard BioScience) using QuantaSmart (v.5.2). Brain tissue sections (thickness, 20 μm) were prepared on a slicing microtome (Leica Microsystems) to perform imaging and observe cerebral localization by digital autoradiography with a high-performance β-Imager (Biospace Lab) using BetaAcquisition software (v.9.4.12. 219) allowing real-time [3H] radioactive imaging through direct β-particle counting in dried tissue sections (detection threshold of 0.007 cpm mm−2).
NOR test on a neurodevelopmental mouse model of schizophrenia induced by PCP administration
Male and female mice used were C57BL/6J mice (Janvier Labs) and were injected subcutaneously with 10 mg per kg of phencyclidine (PCP mice) (Sigma-Aldrich, ANSM authorization, A-2018-3-330-S) or with a saline solution (0.9% (w/v) NaCl) (control mice) at postnatal day 7 (P7), P9 and P11. At adulthood, 1 week before the test, the mice were extensively handled by the operator: the first 2 days, the operator put his hand on the home cage of the animals to familiarize the animals to his presence and, the following days, the animals were handled few minutes per day. NOR testing was carried out in an open field, consisting of four compartments of 50 × 50 cm, each containing one mouse, placed in a dimly lit room with clearly visible contextual cues (black on white patterns) on the surrounding walls. Each mouse was habituated to the arena for 10 min the day before the training session. Mice then performed the NOR task. After a 5-min training session, a 24-h or a 7-day retention interval was observed, during which the mice were transferred back to the home cage and the arena and objects were cleaned. During the test session, the animals were placed back into the arena in the presence of an identical copy of one of the familiar objects and a novel object and allowed to explore the objects freely for 5 min. The objects were plastic toys (approximately 3 cm width, 3 cm length, 5 cm height) and were cleaned with 10% ethanol between sessions. The experiments were video-recorded (EthoVision XT 13.0, Noldus; Logitech Capture v.2.08.11) and exploration times (nose in contact or sniffing at <1 cm) were measured by a blinded observer using a virtual timer (XNote Timer v.1.12.0.0). Mice with a total time exploration of less than 3 s in the test session were excluded. Discrimination indexes ((exploration time of novel object − exploration time of familiar object)/total object exploration time) were compared between groups. For acute treatment, i.p. administration of LY379268 (1 mg per kg) was done 1 h before the training and testing session; i.p. administration of DN13–DN1 (10 mg per kg) was done once 3 h before the training session with the objects; and LY341495 (3 mg per kg) was applied 30 min before DN13–DN1 administration. The second test was performed after a 7-day interval without further compound administration. i.c.v. administration of DN13–DN1 or DN1 (4 pmol in 5 μl) was done 24 h before the training session through a cannula that was implanted 7 days earlier. Cannula implantation was performed on mice anaesthetized with a mixture of ketamine (Imalgene 500, 50 mg ml−1, Merial) and xylazine (Rompun 2%, 20 mg ml−1, Bayer) diluted in 0.9% (w/v) NaCl solution (saline) (2:2:1, i.p., 0.1 ml per 30 g) and mounted onto a stereotaxic apparatus using flat skull coordinates57. Stainless steel guide cannula (22 gauge, 5.00 mm, World Precision Instruments) were implanted in the ventricle (anteroposterior = −0.5 mm; mediolateral = 1.1 mm; dorsoventral = −2.2 mm). Cannulas were implanted vertically in the coronal plane to avoid damage to the wall of the lateral ventricle. The guide cannula was fixed to the skull with anchor screws and dental acrylic (AgnTho’s). Mice could recover for a minimum of 7 days before behavioural testing. Subchronic drug administration consisted of an initial i.p. administration of DN13–DN1 (10 mg per kg) followed by three injections at 1 mg per kg, each injection spaced 1 week apart. The last injection was performed 24 h before the training session and 48 h before the testing session with the objects.
Y-maze test on the genetic mouse model GluN1-KD mice
GluN1-KD mice and their WT littermates’ mice were injected i.p. with LY379268 (1 mg per kg) or DN13–DN1 (20 or 10 mg per kg) 1 h and 3 h before the test, respectively (cohort 1). Independent cohorts of mice were treated with these compounds (LY379268 and DN13–DN1) 7 days before the behavioural test (cohort 2). Subchronic drug administration consisted of an initial i.p. administration of DN13–DN1 (10 mg per kg) followed by three injections at 1 mg per kg, each injection spaced 1 week apart. The last injection was performed 3 h before the behavioural test. Working memory was assessed in a Y-maze as described previously58. The Y-maze consisted of three equivalent arms (38 × 7.6 × 12.7 cm; San Diego Instruments) that were symmetrically disposed at 120° angles from each other. The maze floor and walls were constructed from opaque-white Plexiglas and posters with distinctive geometric shapes (6 × 9 cm) were placed at the end of each arm of the Y-maze. A mouse was initially placed into one arm and the sequence (such as ABCCAB) and the number of arm entries were recorded for 5 min. Results were collected semiautomatically using video-tracking software (Biobserve Viewer v.2.0). Arm entries were defined as all four paws entering the arm. Spontaneous alternation refers to visiting all three arms in sequence (that is, ABC or CAB but not CBC). The percentage of alternations was defined according to the following equation: percentage alternation = [(number of alternations)/(total arm entries − 2)] × 100. The number of arm entries serves as an indicator of ambulation.
PPI of the acoustic startle response test on the genetic mouse model GluN1-KD mice
The test was performed twice on the same cohort of GluN1-KD mice (cohort 3): the first test was performed on the days of LY379268 and DN13–DN1 administration and the second test was performed after a 7-day interval without further compound administration. LY379268 (1 mg per kg) and DN13–DN1 (20 or 10 mg per kg) were administered i.p. only once 1 h and 3 h before the first test, respectively. Subchronic drug administration consisted of an initial i.p. administration of DN13–DN1 (10 mg per kg) followed by three injections at 1 mg per kg, with each injection spaced 1 week apart. The last injection was performed 3 h before the behavioural test. The PPI was measured in four sound-attenuated chambers (SR-LAB ABS System, San Diego Instruments) as described previously58,59 and using SR_LAB Analysis software (San Diego Instruments, 6300-0000-Y). Pulse-only trials consisted of a single white noise burst (120 dB, 40 ms). Prepulse + pulse trials (PP69P, PP72P and PP81P) consisted of a prepulse of white noise (20 ms at 69 dB, 72 dB and 81 dB respectively) that was followed 100 ms after prepulse onset by a startling pulse (120 dB, 40 ms). No-stimulus trials consisted of background noise only (65 dB). The sessions were structured as follows: 1) 5-min acclimatization at background noise level; (2) five pulse trials; (3) ten blocks each containing all eight trials (Pulse, PP69; PP69P, PP72, PP72P, PP81; PP81P, No-Pulse) in pseudorandom order; and (4) five pulse trials. The force intensity for each trial was recorded as the startling level. The percentage of PPI induced by each pre-pulse intensity was calculated as [1 − (startle amplitude on prepulse trial)/(startle amplitude on pulse alone)] × 100%. The same cohort of mice was tested in PPI after 3 h and retested 7 days later as post-drug-treatment intervals, as PPI is not affected by retesting60.
Locomotor activity
For the neurodevelopmental mouse model of schizophrenia, horizontal activity was measured for 60 min in a non-stressful environmental, in a circular corridor (Imetronic). Counts for horizontal activity were incremented by consecutive interruption of two adjacent beams (mice moving through one-quarter of the circular corridor). Locomotor activity was assessed at 3 h after injection of the different nanobodies. For the genetic mouse model GluN1-KD mice, an open-field test was used to assess the locomotor activity as previously described61 under dim light (30 Lux). Test mice were placed into a novel environment—clear Plexiglas chambers measuring 20 × 20 × 45 cm—for 1 h. Their locomotor activity was recorded and tracked by digital activity monitors from Omnitech Electronics through infrared beam sensors. The distance travelled was analysed in 5 min bins to assess locomotor activity. Locomotor activity was assessed 3 h after injection of the different nanobodies.
Inclined platform
A mouse was placed onto a wire platform (1 cm × 1 cm mesh), which was then inclined to 45°. The time it took for a mouse to traverse the entire 24 cm length of the platform was recorded. Mice that fell off the inclined platform were assigned a time of 120 s (ref. 62).
Rotarod
Mice were placed onto an accelerating rotarod (Ugo Basile) and were observed at an initial speed of 4 rpm for 30 s. The rod was then gradually accelerated at a rate of 0.2 rpm s−1. The latency to fall was recorded with a cut-off time of 3 min. Mice were given 3 trials with 10 min rests between trials. The latency to fall for each mouse was counted by averaging over the three trials62.
Pinch-induced catalepsy
Pinch-induced catalepsy was induced as originally described63. A mouse was firmly pinched by the scruff of the neck with a thumb and a forefinger for 5 s. The mouse then was gently placed onto parallel wooden bars (diameter 0.5 cm), with its front paws positioned at 45° angle above the hind paws and elevated for 8 cm above the experimental table. The duration of freezing (catalepsy) was recorded for up to 2 min per trial. Each animal was tested in 3 trials with 2 min intervals. The average duration of catalepsy was analysed.
Brain tissue preparation and cell dissociation
After acute or subchronic treatment, mice were euthanized by cervical dislocation, and the brains were collected and dissected in cold PBS (pH 7.4) as previously described54. For each brain, different regions were dissected: olfactory bulbs, prefrontal cortex, cortex, striatum, hippocampus, cerebellum and midbrain. Samples were collected in a 1.5 ml cryogenic tube (Thermo Fisher Scientific) in 1 ml cold cryopreservation medium (DMEM-GlutaMAX supplemented with 10% FBS and 10% DMSO), frozen at −80 °C in a freezing box and kept at −80 °C until use. On the day of the experiment, the samples were rapidly thawed in a water bath at 37 °C and tissues were washed twice with DMEM-GlutaMAX and twice with cold PBS. Tissues were then digested with 200 μl Versene solution (Thermo Fisher Scientific) for 20 min at 20 °C, and the cells were dissociated by pipetting several times. Several rounds of cell dissociation in DMEM-GlutaMAX were performed to get the maximal number of cells. Dissociated cells were then centrifuged at 3,000g for 5 min, and the cell pellet was washed with 1 ml of cold PBS and resuspended in 200 μl cold PBS.
Relative quantification of mGlu2 expression by TR-FRET
The TR-FRET protocol was performed as previously described54. In brief, 10 μl of dissociated cells were added in a low-volume 96-well microplate (Revvity) and incubated with 10 μl DN10–d2 (25 nM), 10 μl DN1-Tb (25 nM) and 10 μl LY379268 (10 μM), to reach a total volume of 40 μl. Plates were incubated overnight at 22 °C and the sensitized acceptor emission (665 nm) was measured after excitation at 337 nm on the Pherastar FS plate-reader, in the optimal window as previously described64. The resulting acceptor ratio window W2 (50–100 μs)/window W3 (1,200–1,600 μs) multiplied by 104 was then plotted. In parallel, the total amount of proteins was determined using the BCA protein assay (Sigma-Aldrich). Absorbances were measured using the Infinite M200 system.
Animal experiment sample size determination, randomization and blinding
The sample size was determined through previous experience for the neurodevelopmental model65; through power calculations using experimentally identified s.d. values and anticipated meaningful effect sizes for the genetic model66; and using the free application developed by Data’Stat for the in vivo cerebral quantification (http://appsonline.idele.fr/CalculEffectif/). Randomization principle was used to assign experimental mice to treatment groups to avoid bias and spurious conclusions in all of the animal experiments67. In the neurodevelopmental model, all pregnant female mice (Janvier Labs) were not reused to generate new litters. At weaning, each mouse was labelled with a unique ear tag. Within a cage, mice were randomly assigned to the different treatments tested. Thus, mice from different litters received the same treatment for a given cohort. Investigators were blinded to all of the experimental conditions whenever possible during data collection and they were blinded during data analysis. Further details are provided in the Reporting summary.
Statistical analyses
Data are expressed as the mean ± s.d. unless notified otherwise in the figure legend. The main experimental findings are representative of at least two independently performed experiments. The distribution of data in each set of experiments was initially analysed using Shapiro–Wilk’s normality test using GraphPad Prism v.10.4.2 (GraphPad software). When datasets were normally distributed, parametric tests were performed: one-way ANOVA or Brown–Forsythe and Welch followed by post hoc Dunnett’s multiple-comparison test. Behavioural analysis using GluN1-KD mice was analysed using two- or three-way ANOVA with or without repeated measures (rm-MANOVA and MANOVA) where necessary with further post hoc analysis (Tuckey’s HSD test). When datasets were not normally distributed, non-parametric Kruskal–Wallis tests were used followed by a Dunn’s post hoc analysis. P < 0.05 was defined as a significant difference for all statistical analyses. Statistical analyses were performed with GraphPad Prism v.10.4.2 or TIBCO Statistica (v.14.0.0.15, TIBCO).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.