Human sample collection and cell lines
Our study was conducted in accordance with the Helsinki Declaration. Anonymous blood donations from the Charité Campus Mitte blood bank and sputum samples from patients with CF were collected after obtaining informed consent. Both blood and sputum sample collection were approved by the ethics committee of Charité University Hospital, Berlin, Germany. The PLB-985 cell line (female; RRID: CVCL_2162) was donated by M. Dinauer. The PLB-985 cell line was used exclusively as a reference or control, and mycoplasma testing of the cell lines was not performed. The MPO -knockout PLB-985 line was generated in our laboratory45.
Neutrophil isolation and cell culture
Blood was collected into EDTA containing tubes, layered 1:1 on Histopaque 1119 (Sigma-Aldrich) followed by centrifugation for 20 min at 800g. Plasma and the upper layers of the separated blood, consisting mainly of peripheral blood mononuclear cells, were discarded. The neutrophil-rich pink layer was collected and the most dense layer consisting of red blood cells was left undisturbed. Neutrophils were washed in PBS containing 0.1% human serum albumin (HSA, Grifols), and further fractionated on a discontinuous Percoll (Pharmacia) gradient consisting of 2 ml layers with densities of 1,105 g ml−1 (85%), 1,100 g ml−1 (80%), 1,093 g ml−1 (75%), 1,087 g ml−1 (70%) and 1,081 g ml−1 (65%). Neutrophils were carefully layered on the top of the gradient and centrifuged for 20 min at 800g, the interface between the 80% and 85% Percoll layers was collected and washed with PBS containing 0.05% HSA. Neutrophil purity was determined to be >95% by flow cytometry.
NET induction
Primary neutrophils cultured in RPMI (Invitrogen) supplemented with 10 mM HEPES and 0.05% HSA were induced to form NETs using 100 nM phorbol myristate acetate (Sigma-Aldrich), 20 µM nigericin (Sigma-Aldrich), 10 nM Panton–Valentine leukocidin (IBT Bioservices) or 100 µg ml−1 MSU crystals (Sigma-Aldrich) for at least 4 h. NET induction was checked using 1 µM Sytox Green (Thermo Fisher Scientific). After 4 h, >90% of neutrophils had undergone NETosis in all biochemical experiments performed.
Fractionation of NET nucleosomes
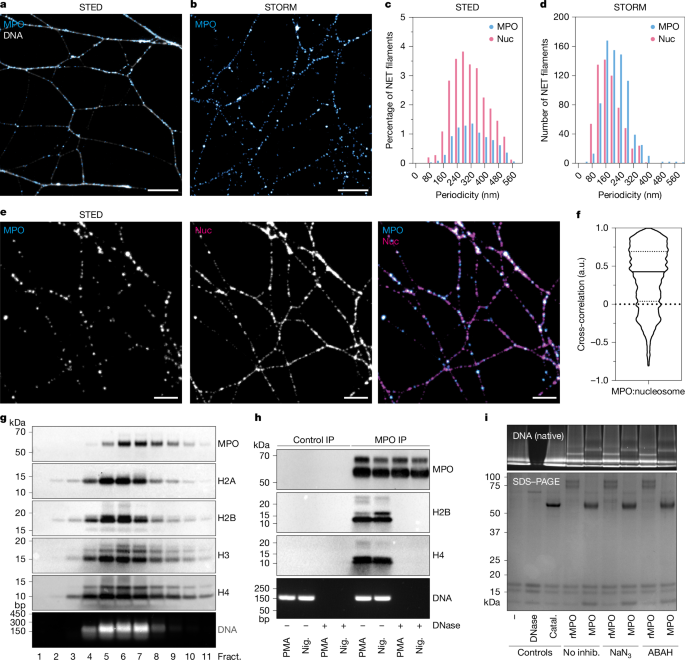
The day before fractionation, continuous 10–30% sucrose gradients (1 mM EDTA, 0.5 mM EGTA, 50 mM KCl, 10% sucrose (w/v) or 30% sucrose (w/v) plus protease inhibitors) were prepared and stored at 4 °C overnight. Then, 2 × 107 neutrophils in 10 ml were plated onto 100 mm × 20 mm Petri dishes (Sarstedt) and then incubated at 37 °C under 5% CO2 for 10 min before the addition of 100 nM PMA. After 4 h, the medium was gently removed, followed by one wash with 10 ml of PBS. After removing the PBS, NETs were digested on a plate with 5 ml of micrococcal nuclease buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2, 1 mM CaCl2) containing 0.2 µl (5.58 U ml−1) micrococcal nuclease (Thermo Fisher Scientific), protease inhibitors, neutrophil elastase inhibitor (Calbiochem) and cathepsin G inhibitor (Merck) for 8 min at room temperature. The digested reaction was stopped by the addition of 5 mM EDTA. After collecting the supernatant containing the digested nucleosomes, the preparation was centrifuged at 3,000g to remove any particulate matter and then transferred to and filtered over 35 ml 100 kDa MWCO columns (Amicon). The retentate was further washed three times in micrococcal nuclease buffer with protease inhibitors and then concentrated to 0.5 ml. The nucleosome preparation was then diluted 1:1 with HEPES pH 7.5, 200 μg ml−1 BSA and 50 mM KCl to correct for osmolality at the interface between the sample and the sucrose gradient. Nucleosomes were then layered on top of 10–30% sucrose gradients and centrifuged in a Beckman Coulter Ultracentrifuge using a SW40 rotor at 36,000 rpm for 18 h at 4 °C. After centrifugation, the gradient was fractionated by careful pipetting from the top of the meniscus. The fractions were then split into two for western blotting or DNA agarose gel analysis.
Protein gels, western blot and native PAGE
Protein preparations for western blotting were reduced in 1× LDS sample buffer (Invitrogen) and DTT was added at a final concentration of 100 mM before boiling at 70 °C for 15 min. The samples were run at 120 V for 1.5 h in MES running buffer (Invitrogen) using the NuPage Invitrogen Mini gel tank system in precast 4–12% gradient Bis-Tris gels. The gels were then directly stained with Instablue (Abcam) or transferred using a BioRad wet tank system onto a 0.22 µm PVDF membrane (Amersham). After transfer, the membranes were blocked for 1 h in 5% BSA followed by primary antibody overnight (Cell Signaling, mouse anti-H2A L88A6, 1:1,000; Abcam, rabbit anti-H2B, ab1790, 1:5, 000; Abcam, rabbit anti-H3, ab1791, 1:5,000; Abcam, rabbit anti-H4, ab10158, 1:5,000; MPO DAKO, A0398, 1:10,000) and then a secondary HRP conjugated antibody (Jackson Labs 1:20,000) for 1 h before washing, and the bands were developed with ECL (Pierce) using the Bio-Rad ChemiDoc. For native-PAGE, MES running buffer was replaced with native running buffer (Invitrogen) and NativePAGE gradient gels (3–12% or 4–16% Bis-Tris gels). NativePAGE sample buffer (Invitrogen) was added to protein samples before loading directly into gels and running in the cold room at 4 °C. Gels were then stained with SybrGold (Invitrogen), ethidium bromide (Sigma-Aldrich), Instablue (Abcam) or Silver stain (Thermo Fisher Scientific).
Super-resolution microscopy
Super-resolution microscopy was performed and analysed using NanoNET as described previously46 using identical microscopes setups. In brief, 1.5 × 105 freshly isolated neutrophils were seeded onto high-precision coverslips (diameter, 24 mm, 1.5H) in six-well cell culture dishes in RPMI supplemented with 0.1% HSA. NET formation was induced by incubation with 100 nM PMA or 20 µM nigericin (Sigma-Aldrich) for 4 h at 37 °C under 5% CO2. The samples were then fixed in 3% paraformaldehyde (PFA) (w/v) (Electron Microscopy Sciences) for 12 min at room temperature. The coverslips were washed twice with PBS. The samples were then blocked in fish gelatin/goat serum blocking buffer for 1 h. The samples were then incubated with primary antibodies in fish gelatin/goat serum blocking buffer at 4 °C overnight (PL2.3, in-house generated (2–5 µg ml−1); 3D9 in-house generated (10 µg ml−1); MPO DAKO, A0398 (1:500)). After two washes with PBS, secondary antibodies (Alexa Fluor (Invitrogen) or CF dyes (Sigma-Aldrich), 1:500) and DNA dyes were added to coverslips in blocking buffer for 1 h. STORM samples were mounted on concave microscopy slides with 100 μl oxygen scavenging buffer (0.1 mg ml−1 GLOX, 0.1 mg ml−1 HRP, 25 mM HEPES, 5% glycerol, 25 mM glucose in PBS, pH 6.0) and sealed with dental imprint adhesive. For SIM and STED microscopy, coverslips were mounted onto microscopy slides using Prolong Gold mounting medium. Auto- and cross-correlograms were generated using NanoNET and plotted using GraphPad PRISM 5. All macros and scripts are available at GitHub (https://github.com/ngimber/NanoNET).
Immunoprecipitation of MPO from sputum samples
Sputum samples were reduced at 37 °C by adding an equal volume of 0.1% DTT in micrococcal nuclease buffer containing protease inhibitors, neutrophil elastase inhibitor and cathepsin G inhibitor for 2 h on a rotating wheel. After 2 h, the samples were vortexed and micrococcal nuclease (30 U ml−1) was added to digest the internucleosomal DNA for 2 h. The reaction was stopped by adding EDTA to a final concentration of 5 mM. After centrifuging the samples at 2,000g for 20 min, the soluble fraction was collected, washed three times and concentrated over 100 kDa MWCO columns followed by fractionation over sucrose cushions. Only mononucleosomes were collected after fractionation on sucrose cushions containing no EDTA or EGTA. DNase I buffer was added to pooled, collected mononucleosome fractions and the sample was then split into two and DNase I (Thermo Fisher Scientific) was added to one of the samples as a control. The samples were then incubated with 2 µg anti-MPO (DAKO) overnight at 4 °C followed by the addition of 20 µl magnetic protein G beads (Invitrogen) for 30 min at room temperature. Magnetic separation of the beads was performed followed by four washes (150 mM NaCl, 1% Triton X-100, 50 mM Tris-HCl pH 7.2, 0.15% BSA, 15% (v/v) glycerol, protease inhibitors), and the beads were then eluted in LDS sample buffer for 15 min at 70 °C. Inputs were calculated as the total amount of DNA per sample after fractionation and before DNase I digests using the Qubit double-stranded DNA quantification assay (Invitrogen). The samples were then western blotted and probed with antibodies against histones or MPO. DNA inputs were monitored and visualized by agarose gel electrophoresis and ethidium bromide staining.
MPO–nucleosome electrophoretic shift assays
HeLA mononucleosomes or recombinant biotinylated nucleosomes (Epicypher, 16-0002 (HeLa mononucleosomes), 16-0006 (recombinant wild type) and 16-0027 (recombinant tailless)) were co-incubated with either recombinant MPO (RnD systems) or native MPO (Sigma-Aldrich) at a molar ratio of 1 nucleosome to 0.5 rMPOs or 0.25 nMPOs for 5 min at room temperature in chromatin remodelling buffer (12 mM HEPES, 40 mM Tris-HCl, 0.32 mM EDTA, 3 mM MgCl2, 10% glycerol, 0.02% Igepal, 60 mM KCl and indicated NaCl concentrations, pH 7.4) and then directly loaded into native gels to monitor binding of MPO to nucleosome after the addition of native sample buffer (Invitrogen). Tris-HCl, NaCl, KCl and MgCl2 were not added to the buffer for saltless conditions. Sodium azide (Roth) and ABAH (Merck) were used as inhibitors of MPO catalytic activity and all stocks were checked for inhibitor activity before aliquoting. DNase I digestion of nucleosomes acted as a fiducial in native gels and catalase was used as a control for a non-nucleosome binding NET protein. For experiments at higher MPO:NUC ratios, incubation times were increased to 10 min. All of the experiments were performed in a 20 μl reaction in which the concentration of nucleosomes was fixed at 500 nM and the MPO molar ratios were calculated accordingly.
Nucleosome pull-down
Biotinylated nucleosomes (Epicypher, 16-0006) were resuspended in 100 μl chromatin remodelling buffer at 1 ng μl−1. rMPO and native MPO were then added to the nucleosomes at different molar ratios. A small aliquot from each reaction was used to monitor inputs. After 20 min, 400 μl of chromatin remodelling buffer containing 40 µl of magnetic streptavidin hydrophilic beads (New England Bioscience) were then added to each sample and the samples were incubated for a further 10 min on a rotating wheel. The samples were then magnetically separated and the post-pull-down supernatants were retained for analysis. The samples were then subjected to four 0.5 ml washes in chromatin remodelling buffer containing 350 mM salt. The post-wash supernatants were also retained and pooled with the post-pull-down lysates. The pooled samples were then desalted in Zeba Spin Columns (Thermo Fisher Scientific) and protein was precipitated on ice for 1 h by adding trichloroacetic acid to a final concentration of 20% (w/v). The precipitated protein was washed three times in ethanol and once in acetone and dissolved in LDS-sample buffer before being boiled at 70 °C for 10 min. The beads were then eluted in LDS sample buffer at 70 °C for 10 min. The samples were then analysed by western blotting and agarose gel DNA electrophoresis.
GATC nucleosome remodelling assay
Chromatin remodelling buffer without EDTA from nucleosome shift assays was used as a reaction buffer. GATC nucleosomes were prepared as a 4× 400 nM stock solution and 5 µl of this preparation was pipetted into a 1.5 ml low-bind reaction tube (Eppendorf). For experiments using the PL2-6 scFv antibody fragment (Creative Biolabs), the scFv fragment was added directly to the nucleosome stock solution at a molar ratio of 3:1 leading to a final 4× stock solution of 1,200 nM PL2-6:400 nM nucleosome, which was incubated at room temperature for 30 min before performing the rest of the experimental protocol. DpnII (NEB) was prepared as a 4× 10 U µl−1 stock solution and 5 µl was added to the GATC nucleosomes. A 2× stock of MPO at molar ratios corresponding to 1:1 or 1:2 nucleosome to MPO (for rMPO this corresponded to 200 nM and 400 nM solutions and for native MPO this corresponded to 100 nM and 200 nM to correct for absolute protein). To start the reaction 10 µl of MPO was added to GATC nucleosomes and to quench the reaction at various timepoints 20 µl of 2× quench buffer (10 mM Tris pH 7.4, 40 mM EDTA, 0.6% SDS and 50 µg ml−1 proteinase K) and the samples were then immediately incubated at 55 °C for 30 min to remove proteins before running DNA on native polyacrylamide gels and visualizing with SybrGOLD. Catalase or horseradish peroxidase was used as a control. The GATC restriction site within the Widom-601 sequence is highlighted in bold in the following sequence: GAACCAATGGGACCATGCTTCACACCGATATCATCGCTTATGTGTTGAATTCATCAGAATCCCGGTGCCGAGGCCGATCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCGATGATGATGGATAGATGGATGATGGATGGATGGATGATGATGGATGAATAGATGGATGGATGAAGCTT.
Sample preparation and cryo-EM data acquisition of recombinant MPO in complex with nucleosomes
Nucleosomes comprising Xenopus laevis H2A, H2B, H3 and H4 histones and Widom-601 145 bp DNA at 4.3 mg ml−1 (~22 µM) were a gift from A. Musacchio, M. Pesenti and D. Vogt; reconstitution was carried out as described previously47. Recombinant MPO was obtained from bio-techne/R&D Systems (cat. no. 3174-MP) and was dissolved at 2.5 mg ml−1 (~31 µM) in PBS. Both were mixed at final concentrations of 1 mg ml−1 nucleosomes (~5 µM) and 2 mg ml−1 rMPO (~25 µM), incubated on ice for 30 min and applied to a Superdex 200 5/150 Increase column (Cytiva), which was connected to an Äkta Micro FPLC system (Cytiva) and equilibrated with 10 mM HEPES pH 7.5, 100 mM NaCl. The peak fractions were pooled and concentrated to 40 µl.
Next, 4 µl of the sample was applied to glow-discharged UltrAuFoil R 1.2/1.3 300 grids (Quantifoil) and plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). Cryo-EM data were acquired on a 200 kV Talos Arctica microscope (Thermo Fisher Scientific) equipped with a field emission gun at a nominal magnification of 120,000x. A total of 4,573 micrograph movies was recorded on a Falcon III camera (Thermo Fisher Scientific) operated in linear mode at a pixel size of 1.21 Å px−1. A total exposure of 56 e− Å−2 was distributed over 40 frames. Details of data acquisition can be found in Supplementary Table 1.
Sample preparation and cryo-EM data acquisition of native MPO in complex with nucleosomes
Native MPO from human leukocytes was purchased from Sigma-Aldrich (cat. no. 475911) and dissolved at 2.5 mg ml−1 (~17 µM) in 10 mM HEPES pH 7.5, 100 mM NaCl. This sample was mixed with H3 601 nucleosomes (see above) at final concentrations of 1.9 mg ml−1 MPO (~13 µM) and 1.1 mg ml−1 nucleosomes (~5.6 µM) and kept on ice.
For the time-course experiment, 4 µl of the sample was directly applied to glow-discharged UltrAuFoil R 1.2/1.3 300 grids (Quantifoil) after 15 s, 2 min, 5 min, 10 min and 20 min, respectively, and plunge-frozen into liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). Data were acquired on a Cs-corrected 300 kV Titan Krios G2 (Thermo Fisher Scientific) equipped with a field emission gun. For each timepoint, ~5,000–6,000 micrograph movies were recorded in super-resolution mode (super-resolution pixel size 0.34 Å px−1) on a K3 camera (Gatan) at a nominal magnification of 105,000×. 53–54 e− Å−2 was distributed over 60 frames, and the slit width of the Bioquantum electron filter (Gatan) was set to 15 eV.
For the long incubation, the sample was kept on ice for 30 min and then applied to a Superdex 200 5/150 Increase column (Cytiva) which was connected to an Äkta Micro FPLC system (Cytiva) and equilibrated with 10 mM HEPES pH 7.5, 100 mM NaCl, analogously to the sample with rMPO (see above). The peak fractions were pooled and concentrated to 40 µl. Then, 4 µl of the sample was applied to glow-discharged UltrAuFoil R 1.2/1.3 300 grids (Quantifoil) and plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). Cryo-EM data were acquired on a 200 kV Talos Arctica microscope (Thermo Fisher Scientific) equipped with a field emission gun at a nominal magnification of 120,000×. A total of 2,723 micrograph movies was recorded on a Falcon III camera (Thermo Fisher Scientific) operated in linear mode at a pixel size of 1.21 Å px−1. A total exposure of 56 e− Å−2 was distributed over 40 frames.
Details of data acquisitions are provided in Supplementary Figs. 9–14 and Supplementary Table 1.
Cryo-EM data processing
Micrograph movies of the sample comprising rMPO and nucleosomes were pre-processed using cryoSPARC live48, including patch motion correction, patch CTF estimation, particle picking using a Gaussian blob and particle extraction. An initial 2D classification was then performed using a subset of 200,000 particles, followed by ab initio reconstruction of three models using 132,381 particles associated to good 2D classes. The three ab initio models and all 5,847,459 extracted particles were then applied to heterogeneous refinement in cryoSPARC48. An initial homogeneous refinement using the 2,029,928 particles assigned to the best-defined class yielded a resolution of 4.0 Å. The particles were then polished and CTF parameters refined in RELION (v.3.1)49. Two more rounds of ab initio modelling and heterogeneous refinement with 3 and 5 classes, respectively, were performed in cryoSPARC using the shiny particles, yielding a final subset of 663,555 particles. Using these, a non-uniform refinement resulted in a reconstruction at 3.76 Å that was sharpened in PHENIX50 by applying a sharpening B-factor of 252.6 Å2. Details about data processing are provided in Supplementary Table 1 and Supplementary Fig. 8.
Processing of the 5 min timepoint of the sample including native MPO and nucleosomes was performed completely in cryoSPARC. The 5,540 micrograph movies were subjected to patch motion correction (involving twofold binning from super-resolution to native pixel size) and patch CTF estimation. In total, 1,380,469 particles were picked using a Gaussian blob picker with a diameter range of 140–200 Å. Particle picks were inspected using the Inspect Particles tool and 975,892 particles with an NCC score of above 0.190 as well as a local power score of between −2,881 and +1,642 were retained. Of these, 786,181 particles were extracted with a box size of 200 × 200 px after twofold binning and subjected to 2D classification with 150 classes. The 59 well-defined classes were used as templates for optimized particle picking, resulting in 3,659,119 particles after picking, 2,441,110 particles after inspection and 2,098,075 twofold binned particles after extraction. These extracted particles were again 2D classified. Ab initio reconstructions with five models were calculated using the 1,328,276 particles assigned to good 2D classes as well as ab initio reconstructions with three models using 769,799 particles assigned to bad 2D classes.
Subsequently, heterogeneous refinement using all extracted particles and a total of 6 ab initio models (four from the particle subset from good 2D classes and two from bad 2D classes) as reference volumes. This heterogeneous refinement yielded two junk classes corresponding to 3.0% and 19.9% of particles, a class of MPO dimers corresponding to 25.7% of all particles (which suffered from severe preferred orientation that precluded any further high-resolution reconstructions), 22.1% of particles corresponding to a free nucleosome class and classes of nucleosomes bound to MPO monomers (23.2% of particles) and dimers (14.2% of particles), respectively. The particle subsets corresponding to free nucleosomes, monomer–nucleosome and dimer–nucleosome, respectively, were separately subjected to non-uniform refinement, followed by reference-based motion correction (all three subsets were combined in the same run). After this, they were separately refined using non-uniform refinement, local CTF refinement and another round of non-uniform refinement. This yielded final global reconstructions at 2.79 Å (nucleosome), 2.89 Å (MPO monomer–nucleosome) and 3.12 Å (MPO dimer–nucleosome), respectively. Details of data processing are provided in Supplementary Table 1 and Supplementary Fig. 11.
In the case of the dimer–nucleosome complex, refinement was finalized by a scheme of particle subtraction and local refinement with either deleting MPO (refinement centred on nucleosome resulting in 3.01 Å reconstruction) or the nucleosome (refinement centred on MPO resulting in 2.98 Å reconstruction) (Supplementary Fig. 15a). Both focused maps were combined using the volume maximum command in UCSF ChimeraX51.
Processing of the datasets corresponding to the 2 min, 10 min and 20 min timepoints was similar to the 5 min dataset. The exact details are provided in Supplementary Figs. 10, 12 and 13 and Supplementary Table 1.
For the 15 s dataset, the initial processing strategy was also similar to the 5 min dataset up to the first heterogeneous refinement (Supplementary Fig. 9). Subsequently, three out of the six classes corresponding to free nucleosome (25.7% of all particles), MPO dimer–nucleosome (17.2% of particles) and a second MPO dimer–nucleosome arrangement (intermediate state of MPO dimer–nucleosome; 15.2% of all particles) were separately subjected to non-uniform refinement, followed by reference-based motion correction (all three subsets were combined in the same run). Next, they were separately refined using non-uniform refinement, local CTF refinement and another round of non-uniform refinement. This yielded final global reconstructions at 3.11 Å (nucleosome), 3.51 Å (MPO dimer–nucleosome) and 3.58 Å (MPO dimer–nucleosome, intermediate state), respectively. Details of data processing are provided in Supplementary Table 1 and Supplementary Fig. 9.
For the MPO dimer–nucleosome intermediate state, refinement was finalized by a scheme of particle subtraction and local refinement with either deleting MPO (refinement centred on nucleosome resulting in 3.52 Å reconstruction) or the nucleosome (refinement centred on MPO resulting in 3.87 Å reconstruction) (Supplementary Fig. 15b). Despite the lower nominal resolution, the reconstruction focused on MPO dimer was much better defined compared with the global refinement. Both focused maps were combined using the ‘volume maximum’ command in UCSF ChimeraX51.
The dataset of native MPO and nucleosomes after 30 min incubation and size-exclusion chromatography was completely refined in cryoSPARC, by initial patch motion correction, patch CTF estimation and extraction of 1,364,624 particles (unbinned, 256 × 256 pixels) that had been picked using the blob picker with a radius of 140–200 Å. 2D classification with 150 classes yielded 45 well-defined classes that were used as templates for optimized picking using the template picker, resulting in 3,728,694 picked and 3,284,273 extracted particles (unbinned, 256 × 256 pixels). In parallel, ab initio reconstructions with four models were calculated using the 596,787 particles assigned to good 2D classes as well as ab initio reconstructions with four models using 767,837 particles assigned to bad 2D classes. Heterogeneous refinement was performed using the template-picked particles and three initial models from the good 2D classes as well as three models from the bad 2D classes. Then, the 765,797 particles associated to the three good classes of the heterogeneous refinement were subjected to another round of ab initio modelling (5 classes) and heterogeneous refinement (4 of the ab initio models). Two out of the four classes, corresponding to free nucleosomes (837,397 particles) and MPO monomer–nucleosome (863,330 particles), were refined by non-uniform refinement to 4.04 Å and 3.94 Å, respectively. See also Supplementary Fig. 14.
The local resolution of all final, deposited reconstructions is shown in Supplementary Fig. 16.
Model building
For the rMPO–nucleosome, a model of rMPO (PDB: 6AZP, chain A)52 as well as a nucleosome model comprising the X. laevis H2A, H2B, H3 and H4 histones and Widom-601 147-bp DNA (PDB 6R1T)53 were rigidly docked into the reconstructed map and manually adjusted in COOT54. The model was optimized by iterative cycles of model adjustment in COOT and real-space refinement in PHENIX50.
For the free-nucleosome model, PDB 6R1T was used as an initial model as well and optimized by manual adjustments in COOT and using real-space refinement in PHENIX. For the MPO dimer–nucleosome complex (main arrangement), this adjusted nucleosome was used as the initial model along with the crystal structure of the MPO dimer (PDB 1MHL)55; again, model was optimized using COOT and PHENIX. Deletion of one MPO monomer (containing heavy and light chain) from this MPO dimer–nucleosome model yielded the initial model for the MPO monomer–nucleosome complex, which was further optimized by COOT and PHENIX. Finally, the MPO dimer–nucleosome intermediate state model was also initiated by placing the refined nucleosome model and PDB 1MHL into the map resulting from combining both focused maps (see above) and finalized in COOT and PHENIX.
Model statistics were calculated using the MOLPROBITY56 implementation in PHENIX and can be found in Supplementary Table 1.
MPO–DNA-binding assay
Nucleosomal DNA from HeLa or recombinant (Widom-601 sequence) mononucleosomes were extracted using the QIAquick PCR purification kit (Qiagen). A serial dilution series of HeLa or Widom-601 nucleosomal DNA (1,600–25 nM) in chromatin remodelling buffer was then performed and rMPO or MPO was added to nucleosomal DNA at a final concentration of 200 nM in a total volume of 10 µl followed by a 30 min incubation at room temperature. Then, 10 µl of 2× native-PAGE sample buffer was added to each sample and 5 µl of each sample was then subjected to agarose DNA electrophoresis. DNA was visualized by ethidium bromide staining.
Negative-stain EM
Purified native MPO (11 µM) was mixed with Widom-601 DNA (3 µM) and incubated for 15 min on ice. The sample was then diluted 60-fold and applied to glow-discharged copper grids coated with 8 nm continuous amorphous carbon. After 1 min incubation, excess sample was removed by blotting, the grid washed three times with Tris-buffered saline and once with 0.75% (w/v) uranyl formate solution before incubation for 1 min with 0.75% (w/v) uranyl formate. After blotting and drying the grid, the sample was analysed in a Tecnai Spirit electron microscope (Thermo Fisher Scientific) operated at 120 kV. Data were recorded at 59,000× magnification on a TemCam F416 camera (TVIPS), resulting in a pixel size of 1.67 Å
Chemical reduction of native MPO and mass photometry of reduced and non-reduced samples
For mass photometry experiments, 100 nM native MPO was incubated at room temperature with 10 mM or 50 mM DTT in a buffer composed of 20 mM HEPES pH 7.5 and 200 mM NaCl. Mass photometry measurements were performed on a Refeyn TwoMP instrument. For this, the samples were diluted 1:10 in a drop of freshly filtered buffer on the instrument’s cover slide after focussing and directly before measuring. Data were analysed using the DiscoverMP software (Refeyn).
In vitro reduced MPO–nucleosome binding assay
Reduced MPO monomers (444 nM) were co-incubated with HeLa mononucleosomes (888 nM) in 500 µl of chromatin remodelling buffer without EDTA for 30 min at room temperature. Then, 0.5 µg of MPO antibody (DAKO) was added to the samples, which were incubated for a further 30 min followed by the addition of 20 µl of protein A/G beads for 20 min. The samples were then washed four times in chromatin remodelling buffer containing 300 mM salt and the beads were then eluted in reducing LDS sample buffer at 70 °C for 10 min before being subjected to western blotting. Nucleosomes were predigested by DNase I as a control.
Cryo-EM sample preparation, data acquisition, and processing and model building of chemically reduced native MPO samples
Native MPO (2.4 mg ml−1, ~16 µM, Sigma-Aldrich) was incubated for 6 h at room temperature in 10 mM HEPES pH 7.5, 100 mM NaCl and 50 mM DTT. The sample was next mixed with reconstituted nucleosomes (see above) at final concentrations of 1.9 mg ml−1 MPO (~13 µM MPO dimers or ~26 µM MPO monomers if complete reduction and dimer dissociation is assumed) and 1.1 mg ml−1 nucleosomes (~5.6 µM), respectively, and kept on ice for times between 15 s and 60 min.
At the indicated timepoints, 4 µl sample was applied to glow-discharged UltrAuFoil R 1.2/1.3 300 grids (Quantifoil) and plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). Cryo-EM data were acquired on a 200 kV Talos Arctica microscope (Thermo Fisher Scientific) equipped with a field emission gun. For each timepoint, around 5,000–6,000 micrograph movies were recorded in counting mode (pixel size 0.68 Å px−1) on a K3 camera (Gatan) at a nominal magnification of 130,000×. 70.3 e− Å−2 were distributed over 60 frames, and the slit width of the Bioquantum electron filter (Gatan) was set to 15 eV. Details of data acquisitions can be found in Supplementary Figs. 17–20 and Supplementary Table 2.
Data processing routes were similar to the data of non-reduced samples (see above) and all steps were performed in cryoSPARC. Three unique molecular assemblies were obtained: nucleosome bound by one and two MPO monomers, respectively, were present in all four timepoints and could be refined to 2.97 Å and 2.95 Å, respectively, in the 2 min timepoint. Nucleosome bound by one MPO monomer and one MPO dimer was present in sufficient number for reconstruction in all but the 2 min timepoint and was refined to 3.16 Å in the 5 min timepoint, followed by focussed refinements on the MPO dimer and the MPO monomer bound to nucleosome, respectively. All details of data processing can be found in Supplementary Figs. 17–21 and Supplementary Table 2.
Sample preparation and cryo-ET data acquisition of PMA-stimulated NETs
For cryo-ET experiments, neutrophils were purified as described above. Thereafter, neutrophils were applied to glow-discharged cryo-EM grids (Quantifoil, holey carbon film, R 2/1 200). Before freezing, neutrophils were treated with 100 nM PMA (Sigma-Aldrich) for at least 4 h to induce NET formation. Neutrophils were plunge-frozen (37 °C, 90% humidity, backblotting 10 s, blotting force 10 and drain time 2 s), using a Vitrobot Mark IV (Thermo Fisher Scientific). Data were acquired on a 300 kV Titan Krios G3 (Thermo Fisher Scientific) system equipped with a K2 and Bioquantum energy filter (energy width 15 eV, Gatan). Tomograms were taken at a nominal magnification of 42,000× (pixel size 3.445 Å) in a dose symmetric scheme57 (tilt range, ±48°; tilt increment, 2°). The total exposure of approximately 100 e− Å−2 was distributed over 49 micrograph movies (8 frames). Details are provided in Supplementary Figs. 6 and 7.
Cryo-ET data processing
Movies of neutrophil extracellular traps were preprocessed in Warp58, including patch motion correction and patch CTF estimation (Supplementary Fig. 7a). After tilt series alignment in IMOD (v.4.11)59, tomograms were reconstructed at binning 4 (pixel size 13.78 Å px−1) in Warp58. For picking, the clustering workflow in TomoTwin was used38. First, tomograms were embedded with the pretrained model. To identify possible protein complexes of interest, the embeddings are projected on a 2D manifold (UMAP) (Supplementary Fig. 7b). By annotating protein complexes in the tomogram, clusters are identified in the UMAP. Here, it is possible to identify only the embeddings of possible targets. To clean the UMAP, the identified clusters were used to recalculate the embeddings and therefore the UMAP. After several rounds of UAMP polishing, three possible clusters, with a total number of 29,242 particles, were identified. After extraction at binning 1 (box size 643, pixel size 3.445 Å px−1) with Warp58, subvolumes were projected in z direction (box size for projection 64 × 64 × 32) for 2D classification in Sphire 1.4 (Supplementary Fig. 7c). Particles of cluster 3 belonging to classes that showed clear nucleosome features were selected (1,548 particles) and finally refined in Relion (v.3.14)49. Refinement reached a resolution of 31 Å (Supplementary Fig. 7d).
Immunofluorescence microscopy of semi-thin CF sputum cryosections
CF sputum samples were fixed in 2% PFA and 0.05% glutaraldehyde, gelatin-embedded and infiltrated with 2.3 M sucrose according to the method described previously60. For immunofluorescence analysis, 200 nm semi-thin sections were cut with a diamond knife at −79 °C with a RMC MTX/CRX cryo-ultramicrotome (Boeckeler Instruments) and transferred to glass coverslips. The sections were blocked with a buffer containing normal donkey serum, BSA and fish gelatin, incubated overnight using antibodies against MPO DAKO A0398 (1:2,000), citrullinated H3 (Abcam, ab5103, 1:500) or 3D9 (1 µg ml−1). During secondary antibody incubations (Alexa Fluor (Invitrogen) or CF dyes (Sigma-Aldrich) 1:500), the sections were counterstained with DNA dyes Sytox (Invitrogen) and or Hoechst 33258 (Invitrogen). The coverslips were mounted onto glass slides with Mowiol (Carl Roth) and analysed using the Leica Thunder widefield microscopy system.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.