Animals
Transgenic male and female mice (8 to 12 weeks of age) were used for behavioural and biochemical experiments. Trpv1-Cre mice (JAX: 017769), Aldh1l1–creERT2 mice (JAX: 031008), Advillin-Cre mice (JAX: 032536), Cx3cr1–creERT2 mice (JAX: 020940), Ai9 mice (JAX: 007909), GCaMP6f mice (JAX: 024105), MitoTag mice (JAX: 032675), and BKS.Cg-Dock7m +/+ Leprdb/J mice (JAX: 000642) were purchased from Jackson Laboratory. Male and female CD1 mice were purchased from Charles River Laboratories. Generation of Myo10 knockout mice (tm1d) and mouse genotyping were described previously31. Mice were maintained at the Duke animal facility. db/db mice aged 12–16 weeks were used. All mouse experiments were approved by the Duke University Institutional Animal Care and Use Committee (IACUC). Mice were housed in an AAALAC-accredited animal facility under a 12 h:12 h light:dark cycle, with food and water provided ad libitum. All animals were housed at 22 ± 1 °C and 30–70 % humidity.
Human DRG tissues
A total of 16 human DRG samples were obtained from donors with and without diabetes (Supplementary Table 1) through the NDRI, under exemption approval from the Duke Institutional Review Board (IRB, Pro00051508). Diabetes was identified on the basis of medical history provided in the NDRI reports.
Reagents
PTX (T7402), Tam (T5648), oligomycin (O4876), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (C2920), rotenone (R8875), antimycin A (A8674), myxothiazol (T5580), CBX (C4790), CytoB (C2743) and Y-27632 (688001) were obtained from Sigma. Pitstop2 (ab120687) was obtained from Abcam. CPZ was obtained from MCE (HY-15640). TTX (1078) was obtained from Tocris. siMyo10 (siRNA ID: 157002), siMYO10 (siRNA ID: 118391) and negative control siRNA (4459405) were obtained from Ambion Life Technology.
Primary cultures from mouse DRG
Mouse DRG neuron culture
Mouse primary DRG neuron cultures were prepared as described44. In brief, DRG were digested with collagenase (0.2 mg ml−1, Sigma, 10103578001) and Dispase-II (3 mg ml−1, Sigma, D4693) for 60 min at 37 °C. The cells were then mechanically dissociated with a pipette, filtered through a 70-µm nylon mesh, and centrifuged at 300g for 5 min. The cells were plated on glass coverslips coated with 0.1 mg ml−1 poly-d-lysine (Thermo, A3890401) and cultured in Neurobasal medium supplemented with 10% FBS, 2% B27 supplement, and 1% penicillin-streptomycin (Neurobasal-supplemented medium) at 37 °C with 5% CO2.
Mouse DRG SGC culture
Mouse DRG SGCs were prepared as previously described15. DRG were enzymatically digested with collagenase and Dispase-II for 90 min at 37 °C. The tissue was then mechanically dissociated using a pipette, filtered through a 70-μm nylon mesh, and centrifuged at 300g for 5 min. Cells were seeded onto 35-mm cell culture dishes (VWR, 10861-656) without poly-d-lysine coating and maintained in Neurobasal-supplemented medium at 37 °C with 5% CO2. On day 3 in vitro (DIV3), non-SGCs—including neurons—were removed by vigorously shaking the culture dishes by hand for 15–20 s, leaving behind a purified adherent layer of SGCs.
Mouse DRG SGC–neuron co-cultures and mitochondrial transfer analysis
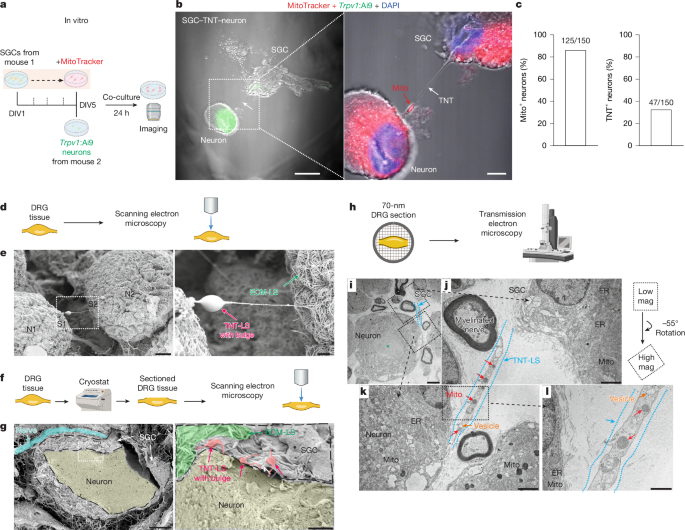
For in vitro mitochondrial transfer experiments, DIV5 SGCs were labelled with 20 nM MitoTracker Deep Red (Thermo Fisher, M22426) in culture medium at 37 °C with 5% CO2 for 30 min. Cells were then washed three times with PBS. MitoTracker-labelled SGCs were collected using 0.25% Trypsin-EDTA (Gibco, 25200-056) and seeded onto coverslips pre-seeded with cultured DRG neurons. Mitochondrial transfer inhibitors, including the TNT formation inhibitor CytoB (3.5 μM) and Y-27632 (10 μM), the endocytosis inhibitor Pitstop 2 (20 μM) or the gap junction blocker CBX (20 μM), were added to the SGC–neuron co-cultures. After 24 h of incubation, cells were imaged using a Zeiss LSM 880 inverted confocal microscope.
Immunocytochemistry in mouse DRG SGC cultures
Primary cultured SGCs were fixed in 4% paraformaldehyde (PFA), then incubated overnight at 4 °C with the primary antibodies: GFAP (mouse, 1:400, Millipore Sigma, G3893), FABP7 (mouse, 1:1,000, Neuromics, MO22188), Kir4.1 (rabbit, 1:500, Alomone, APC-035), AQP4 (rabbit, 1:500, Proteintech, 16473-1-AP), connexin 43 (CX43; rabbit, 1:500, Zymed, 71-0700), glutamine synthetase (GS; rabbit, 1:500, Novus, NB110-41404), and MYO10 (rabbit, 1:500, Sigma, HPA024223). After PBS washes, cells were incubated with appropriate fluorescent secondary antibodies for 1 h at room temperature. Fluorescent images were acquired using a Zeiss LSM 880 confocal microscope.
DRG neuron cultures and assessment of neurite outgrowth
Primary mouse DRG neurons were co-cultured with SGCs for 72 h and subsequently fixed with 4% PFA. Fixed cells were subjected to immunocytochemistry using a βIII-tubulin antibody (1:1,000, Abcam, ab18207) overnight at 4 °C. Immunofluorescence images were acquired with a Nikon fluorescence microscope, and neurite length was quantified using the SNT plugin in ImageJ.
Human DRG SGC–neuron co-cultures and mitochondrial transfer analysis
Postmortem L2–L5 DRG were collected from diabetic and healthy donors and delivered in an ice-cold cell culture medium to the laboratory at Duke University within 24–48 h postmortem. Human DRG cultures were prepared as described45. DRG were digested with collagenase and Dispase-II for 120 to 150 min at 37 °C. Cells were then mechanically dissociated using pipettes and centrifuged (300g for 5 min). Cells were seeded onto poly-d-lysine-coated glass coverslips for neuron culture and into uncoated dishes for SGC culture, and grown in Neurobasal-supplemented medium. On DIV2, non-SGCs were removed by vigorously shaking the culture dishes by hand for 15–20 s. Human SGCs were incubated with 20 nM MitoTracker Deep Red in culture medium at 37 °C with 5% CO2 for 30 min, then co-cultured with human DRG neurons for 24 h.
Whole-mount DRG co-cultured with mouse SGCs and macrophages
Bone marrow-derived macrophages were prepared as described20. In brief, L-929 cells (CCL-1) were obtained from ATCC and authenticated by ATCC. Bone marrow was collected from mice and cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 1% penicillin-streptomycin, and 20% L-929 cell-conditioned medium at 37 °C with 5% CO2. Macrophages or SGCs were seeded on coverslips and labelled with 20 nM MitoTracker Deep Red then co-culture with the whole-mount DRG. CytoB, (3.5 μM) or vehicle was added to medium. Images were captured using a Zeiss LSM 880 inverted confocal microscope.
siRNA transfection in vitro
Transfection was performed using Lipofectamine 2000 (Thermo Fisher, 11668027). In brief, siRNA targeting Myo10, MYO10 and control siRNA was diluted in 100 μl Opti-MEM, and 0.4 μl of Lipofectamine 2000 was diluted in a separate 100 μl Opti-MEM solution. Two solutions were combined and incubated at room temperature for 5 min before being added to the culture dish to make a 15 nM siRNA concentration. The culture medium was replaced 6 h after transfection. siRNA sequences are included as follows: siMyo10: sense, CCUACAAGCAGAGUACAAUtt; antisense, AUUGUACUCUGCUUGUAGGtg; siMYO10: sense: GGUAUUCACUUACAAGCAGtt; antisense: CUGCUUGUAAGUGAAUACCtg.
Mitochondrial health measurement
Mitochondrial membrane potential was assessed using the JC-1 dye (Invitrogen, T3168), a commonly used indicator of mitochondrial health46. Mouse primary SGCs were incubated with JC-1 (2 μg ml−1) for 30 min at 37 °C. JC-1 accumulates in mitochondria in a membrane potential-dependent manner: monomeric forms emit green fluorescence (excitation 485 nm/emission 516 nm), while aggregated forms emit red fluorescence (excitation 579 nm/emission 599 nm), with higher red signal indicating healthier mitochondria. JC-1 fluorescence was monitored in real time using a VivaView FL Incubator Fluorescence Microscope (Olympus) over 24 h. Differential interference contrast images were automatically captured every 30 min at multiple positions.
SIM imaging of mitochondria
Mitochondria of SGCs were labelled with 20 nM MitoTracker Green (Thermo, M7514). SIM imaging was performed using an inverted Zeiss Elyra 7 microscope equipped with a 63× oil-immersion objective. SIM2 image processing was conducted using Zeiss Zen Black software. Mitochondrial length was quantified using the Mitochondria Analyzer plugin in ImageJ (v.1.53q).
OCR measurement
OCR was measured using an XFe96 Seahorse Extracellular Flux Analyzer (Agilent) as described11. In brief, primary DRG neurons and SGCs were seeded in XFe96 cell culture microplates (Agilent, 101085-004) and incubated at 37 °C with 5% CO2 before measurement. On the day of the experiment, cells were washed and placed in Seahorse XF base medium (Agilent, 103575-100) containing 10 mM glucose (Agilent, 103577-100), 1 mM pyruvate (Agilent, 103578-100) and 2 mM glutamine (Agilent, 103579-100). Each assay cycle consisted of 3 min of mixing, 1 min of waiting, and 3 min of OCR measurements. Following three baseline OCR measurements, oligomycin (0.5 μM), FCCP (2 μM), and rotenone/antimycin A (0.5 μM each) were sequentially added. OCR values were normalized to the cell number. Fresh tissue mitochondrial bioenergetics were also measured with the XFe96 Seahorse Extracellular Flux Analyzer as described47. In brief, sciatic nerves or whole-mount DRG were freshly isolated and placed into XFe96 spheroid microplates (Agilent, 102978-100) containing Seahorse XF base medium supplemented with 5.5 mM glucose, 0.5 mM sodium pyruvate and 1 mM glutamine. OCR was normalized to protein content. Oligomycin (12 μM), FCCP (20 μM), and rotenone/antimycin A (20 μM each) were sequentially injected during the assay. An assay cycle of 3 min mixing, 3 min waiting, and 4 min measurement was repeated 3 times for baseline rates and after each port injection.
Calcium imaging in mouse DRG neuron–SGC co-cultures
DRG primary neurons were cultured from Advillin:GCaMP6f mice. Cells were plated on coverslips precoated with poly-D-lysine (Corning, 354087) and grown in a Neurobasal- supplemented medium. Neurons were co-cultured with SGCs following the previously described procedures. The calcium imaging buffer composition was as follows: 140 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, and 10 mM glucose. The buffer solution was perfused through the culture during baseline acquisition, followed by the addition of 100 nM capsaicin, and 60 mM KCl. The calcium indicator Fura2-AM was used for calcium imaging for the TRPV1 antagonist experiment. Dissociated mouse DRG neurons were loaded with 5 μM Fura2-AM (Invitrogen, Thermo Fisher Scientific, F1221) for 45 min and then replaced with calcium imaging buffer. Cells were pretreated with CPZ (100 μM)48 for 3 min prior to capsaicin perfusion. This perfusion procedure was applied uniformly across all four experimental groups. Calcium signals were captured and expressed as F values, representing fluorescence intensity49.
ROS measurement in mouse DRG neuron–SGC co-cultures
ROS levels were measured using the ROS Assay Kit (Thermo Fisher, 88-5930-74) following the manufacturer’s instructions. Primary DRG neuron cultures were assigned to four experimental groups with overnight treatment: (1) vehicle control; (2) PTX (1 μg ml−1) (3) PTX with SGC co-culture; and (4) PTX with SGC co-culture plus CytoB (3.5 μM).
SEM of mouse DRG and spinal cord
For whole-mount mouse DRG and spinal cord imaging, fresh L4–L5 DRG and lumber spinal cord were collected and cut with Vannas spring scissors (FST, 15019-10) to create a flat surface. DRG and spinal cord were fixed in 3% glutaraldehyde in PBS buffer for 1 h at room temperature after tissue collection. After dehydration with 100% ethanol overnight at 4 °C, the samples were ready for the next preparation step. To examine whether trypsin affects the structure of DRG (Extended Data Fig. 3a,b), DRG tissues were treated with 0.25% trypsin for 20 min at 37 °C, followed by hydrolysis with 8 N HCl at room temperature for 20 min, and then fixed in 3% glutaraldehyde, according to a previously described SEM protocol for DRG21. To improve the visualization of TNTs, mice were transcardially perfused with PBS and followed by 4% PFA, and then L4–L5 DRG were collected and immersed in 30% sucrose for over 3 nights at 4 °C. Subsequently, DRG were sectioned by a cryostat (Leica CM 1950). Alternatively, DRG were directly placed in 3% glutaraldehyde for 1 h at room temperature for fixation, followed by dehydration with 100% ethanol overnight at 4 °C. Sample preparation by the critical point drying (Ladd CPD3), then the DRG and spinal cord samples were sputter-coated with gold for 300 s at 12 mA (Denton Desk V). Imaging was performed using an Apreo 2 scanning electron microscope (ThermoFisher Scientific) with a 2.00 kV accelerating voltage and 25 pA emission current at Duke University Shared Materials Instrumentation Facility. Multiple detectors (ETD, T1, T2 and T3) were utilized, and immersion mode with T1, T2 and T3 detectors was used to capture high-magnification images.
SEM and immunostaining of human DRG
Fresh human DRG (L2–L5) were fixed overnight in 4% paraformaldehyde upon delivery, and then immersed in 30% sucrose for at least 3 nights at 4 °C. Free-floating sections (30 μm) were cut using a cryostat. Some sections were processed for SEM, while adjacent sections were used for immunohistochemistry (IHC). For SEM, imaging was performed using an Apreo 2 scanning electron microscope. Multiple detectors (ETD, T1, T2, and T3) were used. For immunohistochemistry, adjacent sections were blocked with 5% bovine serum albumin (BSA) for 1 h at room temperature, followed by overnight incubation at 4 °C with anti-FABP7 antibody (rabbit, 1:100). The next day, sections were incubated with Alexa Fluor 488-conjugated anti-rabbit secondary antibody (1:400) and Nissl/NeuroTrace-640 for 1 h at room temperature. Sections were mounted using DAPI Fluoromount-G and imaged with a Zeiss LSM 780 confocal microscope.
TEM of mouse DRG
Dissected DRG tissue was fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for at least 1 h at room temperature, then stored at 4 °C. Samples were washed with 0.1 M sodium cacodylate buffer, followed by post-fixation in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer (pH 7.2) for 2 h at room temperature. After three additional rinses in buffer, tissues were dehydrated through a graded acetone series and embedded in epoxy resin. Semi-thin sections (500 nm) and ultrathin sections (70–80 nm) were cut using a LEICA EM UC7 ultramicrotome (Leica). Ultrathin sections were collected on copper grids, post-stained with uranyl acetate and lead citrate, and imaged using a JEOL 2100 Transmission Electron Microscope (Duke Center for Electron Microscopy and Nanoscale Technology) at an accelerating voltage of 120 kV.
Animal models of neuropathic pain and inflammatory pain
CIPN was induced by intraperitoneal injections of PTX at 2 mg kg−1 every other day for 4 doses. Type 2 DPN was modelled using db/db mice (JAX: 000642). Type 1 diabetes was induced by intraperitoneal administration of STZ (150 mg kg−1; Sigma-Aldrich, S0130)50. SNI surgery was performed as previously described51. In brief, the common peroneal and tibial nerves were ligated and transected, while the sural nerve was left intact. Inflammatory pain was induced by intraplantar injection of 20 μl CFA into the hindpaw52.
Nerve blockade by bio-resorbable PEU films for controlled bupivacaine release
We previously utilized bio-resorbable PEU films for the controlled release of local anaesthetics in pain management26,53. Bupivacaine-loaded films (40% w/w, ~0.3 mg bupivacaine per film) and blank control films were prepared in the laboratory of M.L.B. Bupivacaine-containing films were wrapped around the ipsilateral sciatic nerve during SNI surgery.
Mitochondrial imaging in the DRG and spinal cord of MitoTag mice
MitoTag mice were crossed with Aldh1l1-creERT2, Advillin-cre and Cx3cr1-creERT2 lines. In inducible Cre lines, recombination was induced by Tam administration54. Tam was prepared in a solution of 10% ethanol and 90% corn oil (Sigma, C8267). Mice received intraperitoneal injections of Tam at 100 mg kg−1 once daily for 5 consecutive days. DRG and spinal cord tissues were collected for mitochondrial imaging analysis.
Mitochondrial isolation and immunocapture in Aldh1l1-MitoTag mice DRG
Mitochondria were isolated from fresh dissected DRG tissue as described23. In brief, fresh DRG were dissected from Aldh1l1-MitoTag mice. The samples were then subjected to nitrogen cavitation, and after depressurization, nuclei and debris were removed by centrifugation. The supernatant was filtered through a pre-separation filter (130-041-407; Miltenyi Biotec) to obtain the crude mitochondrial fraction. For immunocapture of mitochondria against eGFP, the crude mitochondrial fraction was incubated with GFP beads for 1 h. LS columns (Miltenyi Biotec) were then used to separate GFP microbead-coated mitochondria (immunoprecipitate) and mitochondria without GFP (supernatant) from the solution. For western blot analysis, GFP antibodies (goat, 1:1,000, Abcam, ab5450) were used to examine the eGFP, and anti-COX4 (rabbit, 1:1,000, Abcam, ab16056) was used as a mitochondrial loading control.
Generation of AAV-MaCPNS2-Syn-jRGECO1a for DRG neuron targeting
AAV production was performed using the following plasmids: pAdDeltaF6 (Addgene #112867), pUCmini-iCAP-AAV.MaCPNS2 (Addgene #185137), and pAAV.Syn.NES-jRGECO1a.WPRE.SV40 (Addgene #100854). HEK293T cells (CRL-3216) obtained from ATCC, and authentication was performed by ATCC through morphological and STR profiling. HEK293T cells were not tested for mycoplasma contamination. HEK293T cells were cultured in DMEM medium for transfection. Cells were transfected with 30 μg pAdDeltaF6, 15 μg pUCmini-iCAP-AAV.MaCPNS2, and 15 μg jRGECO1a plasmids using PEI MAX (Polysciences, 24765). After 72 h, cells were collected and lysed in 4 ml lysis buffer (15 mM NaCl, 5 mM Tris-HCl, pH 8.5) through three freeze-thaw cycles. Lysates were incubated with benzonase (50 U ml−1; Millipore, 70664) for 30 min at 37 °C and then centrifuged at 4,500 rpm for 30 min at 4 °C. The supernatant was layered onto a stepwise iodixanol gradient (15%, 25%, 40% and 60%) and centrifuged at 67,000 rpm for 1.5 h at 18 °C using a Beckman Ti-70 rotor. The viral fraction was collected from the 40%–60% interface, and concentrated using a 100 kDa molecular weight cutoff filter (Millipore, UFC910008). Purified viral aliquots were stored at –80 °C until use.
Simultaneous ex vivo calcium and mitochondrial imaging in DRG neurons from MitoTag mice
AAV-MaCPNS2-Syn-jRGECO1a (3 × 1011 viral genomes per mouse) was administered intraperitoneally to MitoTag mice at postnatal day 0. Four weeks post-injection, mice underwent SNI surgery and received Tam (100 mg kg−1, intraperitoneal injection) once daily for 5 consecutive days. Ex vivo calcium imaging was performed 3–7 days after surgery. L4–L6 DRG were dissected and incubated in artificial cerebrospinal fluid. Live imaging was conducted using a Zeiss LSM 780 upright confocal microscope equipped with a 20× water-immersion objective. Images were acquired with 50 cycles over a 15 min time-lapse session for each DRG. Data were analysed using FIJI (ImageJ) software.
Intra-ganglionic (DRG) injection of siRNA
siMyo10 and siCtrl were mixed with RVG-9R peptide in 5% glucose at a peptide/siRNA molar ratio of 2:1 before use, following a previously described protocol55. Intra-DRG injection was performed as described previously56. In brief, a partial laminectomy was performed to expose the left L4 and L5 DRG. A total volume of 1 μl containing 4 μg of siRNA was delivered to each DRG using a gelatin sponge as the delivery matrix.
Intrathecal injection
For intrathecal drug delivery, antimycin A (110 ng), CytoB (70 ng and 350 ng), or Pitstop2 (120 ng) was diluted in 10 μl of PBS and administered via lumbar puncture using a 30-gauge needle between the L5–L6 vertebral levels. Successful intrathecal delivery was confirmed by observing a characteristic tail-flick response57.
In vivo labelling of mitochondria in the DRG
Mitochondrial labelling in the DRG was performed based on a previously described protocol with minor modifications58. A total volume of 0.6 μl Mitotracker Deep Red FM (2 μM; Thermo, M22426), with or without CytoB, was injected into the L4–L5 DRG using a Hamilton syringe connected to a glass micropipette. Injected DRG were collected 24 h post-injection, embedded in OCT medium, and sectioned at 20 μm thickness using a cryostat. Immunohistochemistry was then performed on these sections using antibodies against GFAP, along with Nissl staining.
Adoptive transfer of SGCs to mice
SGCs were collected from primary cell cultures, and 8,000 cells suspended in 1 μl PBS were injected intra-ganglionically into the L4 and L5 DRG using a Hamilton syringe connected to a glass micropipette. In select experiments, SGCs were pre-labelled with 20 nM MitoTracker Deep Red FM in culture medium for 30 min at 37 °C, followed by three PBS washes prior to injection.
Mitochondria isolation and adoptive transfer of mitochondria via intra-ganglionic injection
Mitochondria were isolated from SGCs following a previously described protocol59 with minor modifications. In brief, SGCs were detached from culture dishes, pelleted, and resuspended in MIB buffer (210 mM D-mannitol, 70 mM sucrose, 5 mM HEPES, 1 mM EGTA, and 0.5% (w/v) fatty acid-free BSA, pH 7.2) at 4 °C. Cells were homogenized with 25 strokes and centrifuged at 600g for 5 min at 4 °C. The supernatant was collected and centrifuged at 7,000g for 10 min at 4 °C. Mitochondria isolated from 10,000 SGCs in 1 μl of PBS were injected intra-ganglionically into the L4 and L5 DRG using a Hamilton syringe and glass micropipette. In select experiments, to inhibit mitochondrial oxidative phosphorylation, SGCs were pretreated with 1 mM myxothiazol (Sigma, T5580) for 10 min prior to mitochondrial isolation.
Behavioural tests for pain in mice
For von Frey testing, mechanical sensitivity was assessed with a set of von Frey filaments (Stoelting) with logarithmically increasing stiffness ranging from 0.02 g to 2.56 g, which were applied to the hindpaw plantar surface, and a quick withdrawal or licking response to the stimulus was considered a positive response. PWT was calculated using the up-down method. For assessing mechanical allodynia, paw withdrawal frequency (PWF) to a 0.16 g von Frey filament was measured by observing reflexive withdrawal responses within 10 stimulations, and a quick withdrawal or licking response to the stimulus was considered a positive response. Hargreaves test was used to assess heat sensitivity. Paw withdrawal latency was measured with a Hargreaves radiant heat apparatus (IITC Life Science). CPP assay was used to measure ongoing pain in mice44. Mice were habituated for 3 days with 30 min of preconditioning in a two-compartment CPP chamber consisting of white and dark chambers. The baseline behaviour of the mice was recorded on the fourth day using a camera and automatically tracked for 15 min with ANY-Maze software (Stoelting). On the fifth day (conditioning day), mice underwent a 30 min pairing session without intra-DRG injection in the preferred CPP chamber in the morning session, followed by a 30 min pairing session with intra-DRG injection in the non-preferred CPP chamber in the afternoon session. Twenty-four hours later, mice were placed in the CPP test box with access to both chambers, and their behaviour was recorded for 15 min. The time spent in both chambers was analysed using ANY-Maze software. The CPP score, measured in seconds, was calculated as the inverse of the time spent in the preferred chamber, using the formula: post-preference time minus pre-preference time.
Immunohistochemistry in mouse DRG, spinal cord, nerve, and skin tissues
Following terminal anaesthesia with isoflurane, mice were transcardially perfused with PBS, followed by 20 ml of 4% PFA in PBS. The DRG, spinal nerves and hindpaw skins were collected, fixed in 4% PFA and immersed in 30% sucrose at 4 °C for at least 3 nights. Tissues were then embedded in OCT medium (Tissue-Tek), and sections were prepared using a cryostat at the following thicknesses: DRG (20 μm), spinal cord (30 μm), spinal nerve (20 μm), and skin tissue (25 μm). Tissue sections were then blocked in a solution containing 5% BSA and 0.3% Triton X-100 for 1 h at room temperature. Next, sections were incubated overnight at 4 °C with the following primary antibodies: anti-FABP7 (mouse, 1:1,000, Neuromics, MO22188), anti-Iba1 (rabbit, 1:800, Wako, 019-19741), anti-GFAP (mouse, 1:400, Sigma, G3893), anti-MYO10 (rabbit, 1:500, Sigma, HPA024223), anti-TOM20 (rabbit, 1:600, Proteintech, 11802-1-AP) and anti-PGP9.5 (rabbit, 1:200, Thermo, PA5-29012). Following several washes with PBS, the sections were incubated with species-specific secondary antibodies and Nissl/NeuroTracer-640 (1:100, Thermo, N21483). Finally, the sections were mounted with coverslips using DAPI Fluoromount-G mounting medium (Southern Biotech, 0100-20). Images were acquired using a Zeiss LSM 780 confocal microscope. For quantification of immunofluorescence staining, the same acquisition settings were applied to images requiring comparison under different conditions. Three sections were randomly selected from each mouse, and the integrated density of the fluorescence signal per section was measured using ImageJ software (v.1.53q). For intraepidermal nerve IENF analysis in mouse hindpaw skins, all ascending nerve fibre branches crossing into the epidermis were counted. Three randomly selected skin sections were analysed for each mouse.
In situ hybridization and immunohistochemistry in human DRG
Fresh human DRG were immediately fixed in 4% paraformaldehyde overnight at 4 °C upon delivery. Free-floating sections (30 µm) were then cut using a cryostat. For MYO10 in situ hybridization, a human MYO10 RNAscope probe (RNAscope Probe Hs-MYO10, 440691), designed by Advanced Cell Diagnostics was used. The RNAscope protocol was followed according to the manufacturer’s instructions. Following the RNAscope steps, immunohistochemistry was performed as described below. The sections were blocked with 5% BSA for 1 h at room temperature and then incubated with anti-FABP7 antibody (rabbit, 1:100, Thermo, PA5-24949) overnight at 4 °C. Images were captured using a Zeiss LSM 780 confocal microscope with consistent acquisition settings across different samples. The integrated density of the fluorescence signal per section was measured using Image J (v.1.53q).
snRNA-seq and data analysis in human DRG
Human DRG tissues were snap-frozen and stored at –80 °C. Nuclei isolation was performed as previously described60. Single-nucleus capture was performed using the 10x Genomics Chromium Single Cell 3′ system (v.3.1). Libraries from individual nuclei samples were pooled and sequenced on an MGISEQ-2000 platform. snRNA-seq reads were processed using Cell Ranger v.4.0.061, and alignment to the Human GRCh38 (GENCODE v.32/Ensembl98) reference genome performed using default parameters. Downstream analysis was conducted with Seurat v.4.3.062 using R v.4.3.0. Cells were filtered out if they expressed fewer than 200 genes or had more than 10% mitochondrial reads. Gene expression was then normalized and scaled using the NormalizeData and ScaleData functions from Seurat, respectively, with default settings, as well as the FindVariableFeatures function to identify the highly variable genes. Principal component analysis was carried out, and downstream analyses were based on the top 20 principal components. To address batch effects, datasets were integrated, re-normalized, scaled, and batch-corrected using Harmony v.0.1.163. Unsupervised clustering was done using the FindClusters function from Seurat with a resolution of 0.8. UMAP was applied to reduce the data to two dimensions. FindMarkers with default settings to get gene markers, and cell types within each cluster were annotated based on known marker genes and genes that were differentially expressed within each cluster.
Statistical analysis
Data are expressed as mean ± s.e.m. Statistical analyses were completed with Prism GraphPad 8.4. The sample sizes were based on our previous studies44,45. Each data point corresponds to an individual animal. All data were included in the analyses (no outliers removed). Data were analysed using unpaired t-test (two-sided) or Mann–Whitney test (two-sided) for comparison between two groups, one-way ANOVA followed by Tukey post hoc test, and two-way ANOVA followed by Sidak’s multiple comparisons test for two groups and Tukey’s multiple comparisons test for more than two groups. A significance level of P < 0.05 was considered statistically significant. Additional statistical details are included in Supplementary Table 4.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.