Drosophila strains and culture
Flies were grown on media of cornmeal (62.5 g l−1), inactive yeast powder (25 g l−1), agar (6.75 g l−1), molasses (37.5 ml l−1), propionic acid (4.2 ml l−1), tegosept (1.4 g l−1) and ethanol (7 ml l−1) under a 12 h light–12 h dark cycle at 25 °C in approximately 60% relative humidity, unless stated otherwise. To prevent the undesired activation of optogenetic actuators or the photoconversion of all-trans retinal by ambient light, flies expressing CsChrimson or mito-dR and their controls were reared and housed in constant darkness and transferred to food supplemented with 2 mM all-trans retinal (Molekula) in dimethylsulfoxide (DMSO), or to DMSO vehicle only, 2 days before the optical stimulation experiments, at an age of 1–2 days post eclosion. Flies expressing TrpA1 and their controls were cultured and maintained at 21 °C and shifted to 29 °C for 12 h.
Transgene expression was directed to dFBNs by driver lines R23E10-GAL4 (ref. 58) or R23E10 ∩ VGlut-GAL4 (ref. 2) (created by reconstituting GAL4 from hemidrivers59 R23E10-DBD60 and VGlut-p65AD61); projection neurons, Kenyon cells and dopaminergic neurons were targeted by GH146–GAL4 (ref. 62), OK107-GAL4 (ref. 63) and TH-LexA64, respectively. Effector transgenes encoded fluorescent markers for flow cytometry (UAS-6xEGFP65), visually guided patch-clamp recordings (UAS-mCD8::GFP66), mitochondrial morphometry (UAS-mito-GFP67,68) or ratiometric imaging (UAS-RFP or UAS-tdTomato); integral proteins of the outer mitochondrial membrane69 (UAS-OMM-mCherry), endoplasmic reticulum70 (UAS-tdTomato-Sec61β) or plasma membrane71 (UAS-CD4-tdTomato); the ATP sensors iATPSnFR1.0 (refs. 72,73) or ATeam1.03NL74,75; the mitochondrial alternative oxidase AOX of Ciona intestinalis76; delta-rhodopsin of Haloterrigena turkmenica with an IMM-targeting sequence16,77 (mito-dR); the opto- or thermogenetic actuators78 CsChrimson79 or TrpA1 (the latter in UAS- and lexAop-driven versions80,81); the mitochondria–endoplasmic reticulum contact site82,83 or mitophagy84,85 reporters SPLICSshort or mito-QC; overexpression constructs encoding Ucp4A or Ucp4C86,87, Drp1 (3 independent transgenes88,89,90), Opa1 (ref. 89) or Marf (2 independent transgenes89,91); or RNAi transgenes for interference with the expression of Drp1 (ref. 91), Opa1 (7 independent transgenes88,92,93), Marf (5 independent transgenes88,92,93), zucchini93 or Mitoguardin (2 independent transgenes92). Recombinant strains carrying the UAS-Marf and R23E10-GAL4 or R23E10-DBD transgenes on the third chromosome were generated to enable the co-expression of Opa1 and Marf. Endogenous Drp1 was colocalized with mitochondria in Drp1::FLAG-FlAsH-HA flies94. Supplementary Table 3 lists all fly strains and their sources.
Sleep measurements and sleep deprivation
In standard sleep assays, females aged 2–4 days were individually inserted into 65-mm glass tubes containing food reservoirs, loaded into the Trikinetics Drosophila Activity Monitor system and housed under 12 h light–12 h dark conditions at 25 °C in 60% relative humidity. Flies were allowed to habituate for 1 day before sleep—classified as periods of inactivity lasting more than 5 min95,96 (Sleep and Circadian Analysis MATLAB Program97)—was averaged over two consecutive recording days. Immobile flies (fewer than two beam breaks per 24 h) were manually excluded.
Our standard method of sleep deprivation used the sleep-nullifying apparatus98: a spring-loaded platform stacked with Trikinetics monitors was slowly tilted by an electric motor, released and allowed to snap back to its original position. The mechanical cycles lasted 10 s and were repeated continuously for 12 h, beginning at zeitgeber time 12. Flies expressing TrpA1 in dFBNs (and relevant controls) were mechanically sleep-deprived at 29 °C.
To guard against potential side effects of regular mechanical agitation, we explored two alternative methods of sleep deprivation, with comparable results. In some behavioural experiments (Extended Data Fig. 8f), an Ohaus Vortex Mixer stacked with Trikinetics monitors produced horizontal circular motion stimuli with a radius of about 1 cm at 25 Hz for 2 s; in contrast to the rhythmic displacement of flies in the sleep-nullifying apparatus98, stimulation periods were randomly normally distributed in 20-s bins, hindering adaptation. In some morphometric experiments, flies expressed TrpA1 in arousing dopaminergic neurons14, whose activation at 29 °C produced sleep deprivation without sensory stimulation (Extended Data Fig. 5c,d).
Rebound sleep was measured in the 24-h window after deprivation. Cumulative sleep loss was calculated for each individual by comparing the percentage of sleep lost during overnight sleep deprivation with the immediately preceding unperturbed night. Individual sleep regained was quantified by normalizing the difference in sleep amount between the rebound and baseline days to baseline sleep. Only flies losing more than 95% of baseline sleep were included in the analysis.
Arousal thresholds were determined by applying horizontal circular motion stimuli with a radius of ~1 cm at 8 Hz, generated by a Talboys Multi-Tube Vortexer. Stimuli lasting 0.5–20 s were delivered once every hour between zeitgeber times 0 and 24, and the percentages of sleeping flies (if any) awakened within 1 min of each stimulation episode were quantified.
Sleep induction by photo-energized mitochondria
For experiments with photo-energized mitochondria16,77, 3–5-day-old females expressing R23E10 ∩ VGlut-GAL4– or R23E10-GAL4-driven mito-dR and their parental controls were reared for 2 days on standard food supplemented with all-trans retinal (or DMSO vehicle only, as indicated) and individually transferred to the wells of a flat-bottom 96-well plate. Each well contained 150 µl of sucrose food (5% sucrose, 1% agar) with or without 2 mM all-trans retinal. The plate was sealed with a perforated transparent lid, inserted into a Zantiks MWP Z2S unit operated at 25 °C and illuminated from below by infrared LEDs while a camera captured 31.25 frames per s from above. Zantiks software extracted time series of individual movements, which were converted to sleep measurements with the help of a custom MATLAB script detecting continuous stretches of zero-speed bins lasting more than 5 min. After flies had been allowed to habituate for at least one day in the absence of stimulation light, high-power LEDs running on an 80% duty cycle at 2 Hz (PWM ZK-PP2K) delivered about 7 mW cm−2 of 530-nm light for 1 h. Movement was monitored for 24 h, including the initial hour of optogenetic stimulation. If flies were found dead at the end of the experiment, data from the 1-h time bin preceding the onset of continuous immobility (>98% zero-speed bins during two or more consecutive hours until the end of the recording) onward were excluded. The >98% threshold was applied to avoid scoring rare instances of video tracker noise as movement.
Brain dissociation and cell collection for scRNA-seq
On each experimental day, averages of 186 rested and 144 sleep-deprived female flies were retrieved alive from Trikinetics monitors (Extended Data Fig. 1a) and dissected in parallel in ice-cold Ca2+– and Mg2+-free Dulbecco’s PBS (Thermo Fisher) supplemented with 50 µM d(-)−2-amino-5-phosphonovaleric acid, 20 µM 6,7-dinitroquinoxaline-2-3-dione and 100 nM tetrodotoxin (tDPBS) to block excitatory glutamate receptors and voltage-gated sodium channels. The lower number of sleep-deprived flies recovered reflects accidental mortality associated with the operation of the sleep-nullifying apparatus. Brains were transferred to Protein LoBind microcentrifuge tubes containing ice-cold Schneider’s medium supplemented with the same toxins (tSM), washed once with 1 ml of tSM, incubated in tSM containing 1.11 mg ml−1 papain and 1.11 mg ml−1 collagenase I for 30 min at room temperature, washed again with tSM and subsequently triturated with flame-rounded 200-μl pipette tips. Dissociated brain cells were collected by low-speed centrifugation (2,000 rpm, 3 min), resuspended in 1 ml tDPBS, and filtered through a 20-μm CellTrics strainer. For the isolation of cells by flow cytometry, dead cells were excluded with the help of a DAPI viability dye (1 μg ml−1; BD Pharmingen). Single cells were gated for on the basis of forward and side scatter parameters, followed by subsequent gating for EGFP-positive and EGFP-negative cells, using the integrated fluorescence excited at 488 nm and collected in the 500–526-nm band (Becton Dickinson FACSDiva software). Both the EGFP-positive fraction and the EGFP-negative flow-through were collected for sequencing. Samples were partitioned into single cells and barcoded using droplet microfluidics30 (10X Chromium v.3 and v.3.1) and multiplexed during Illumina NovaSeq6000 sequencing. Brains and dissociated cells were kept on ice or at ice-cold temperatures from dissection to sample submission, including during flow cytometry, but not during the enzymatic and mechanical dissociation steps, which took place at room temperature.
scRNA-seq data processing and alignment
Raw transcriptomic data were pre-processed with a custom command line script30,99,100, which extracted cell barcodes and aligned associated reads to a combination of the Drosophila melanogaster genome release BDGP6.22 and the reference sequences of the GAL4 and EGFP-p103’UTR transgenes, using STAR 2.6.1b with default settings101. Flybase version FB2018_03 gene names were used for annotation. The cumulative fraction of reads as a function of cell barcodes, arranged in descending order of the number of reads, was inspected, and only cells with a high number of reads, up to a clearly visible shoulder, were retained30; beads with few reads, potentially ambient RNA, were discarded. All subsequent analyses were performed in R, using the Seurat v.4.1 package102.
Three biological replicates, collected on different days from independent genetic crosses, were merged, variation driven by individual batches was regressed out, and the data were normalized by dividing by the number of unique molecular identifiers per cell and multiplying by 10,000. Applying standard criteria for fly neurons28,100,103,104, we rejected genes detected in fewer than three cells and retained only cells associated with 800–10,000 unique molecular identifiers and 200–5,000 transcripts.
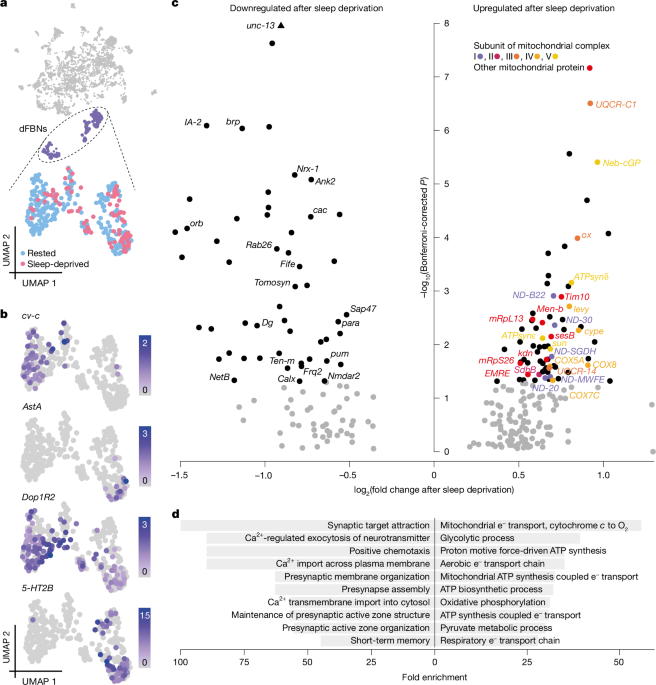
Principal component analysis was used to compress the expression data from an initial dimensionality of 10,000 (the number of variable features) to 50 (the number of principal components we chose to consider); the scores along these 50 dimensions were then visualized in a two-dimensional uniform manifold approximation and projection embedding. Clusters were identified by constructing a shared nearest neighbour graph and applying the Louvain algorithm with resolution 0.2. Clusters were annotated manually according to the presence of established markers100,103: cholinergic, glutamatergic and GABAergic neurons expressed elav and nSyb and, respectively, genes encoding VAChT or the glutamate transporter (VGlut) or glutamic acid decarboxylase 1 (Gad1) at levels >2; Kenyon cells were identified by the expression of eyeless and Dop1R2 and partitioned into αβ, α′β′ and γ divisions according to the distributions of sNPF, fasciclin 2 and trio; monoaminergic neurons were identified by the presence of the vesicular monoamine transporter Vmat and divided into dopaminergic, serotonergic and octopaminergic–tyraminergic neurons by the co-expression of genes encoding biosynthetic enzymes (dopa decarboxylase, tyrosine hydroxylase, tyrosine decarboxylase 2, tryptophan hydroxylase and tyramine β-hydroxylase) and vesicular transporters for dopamine or serotonin (DAT and SerT); projection neurons were defined and classified by the expression of the transcription factors cut and abnormal chemosensory jump 6 (acj6), with or without Lim1; glia lacked elav and nSyb but expressed the Na+–K+ ATPase encoded by the nervana 2 gene, whereas astrocytes also contained the astrocytic leucine-rich repeat molecule (alrm); cells of the fat body were recognized by the expression of Secreted protein, acidic, cysteine-rich (SPARC), Metallothionein A (MtnA), I’m not dead yet (Indy) and pudgy; R23E10 neurons expressed elav and nSyb plus AstAR-1 and EGFP at a level >2. Expression level cutoffs for VAChT, VGlut, Gad1 and EGFP were chosen to bisect bimodal distributions (Extended Data Fig. 1e). For reclustering neurons expressing specific fast-acting neurotransmitters, dFBNs, Kenyon cells or projection neurons, 150 genes were used as variable features.
Differential gene expression and gene ontology analyses
Differentially expressed genes were identified in Seurat by means of the ‘FindMarkers’ function, using the ‘RNA assay’ counts of the two comparison groups, and restricted to genes detected in ≥1% of cells in either of the two groups with, on average, a ≥0.01-fold (log scale) expression level difference between the rested and sleep-deprived states. Expression levels were compared by Bonferroni-corrected Wilcoxon rank-sum test.
Gene ontology terms enriched in the set of differentially expressed nuclear genes were identified using PANTHER v.17 or the ViSEAGO v.1.4.0 and topGO v.2.42.0 packages. PANTHER compared the list of differentially expressed genes with the Drosophila melanogaster reference list and the gene ontology annotation database (https://doi.org/10.5281/zenodo.7942786, version 2023-05-10). In ViSEAGO and topGO, differentially expressed genes were compared to a reference set of all variable genes used in Seurat and annotated in Ensembl; only gene ontology terms with more than 40 attached genes were considered, and enriched terms with unadjusted P < 0.001 were clustered hierarchically according to Wang’s distance105.
Two-photon imaging
Females aged 3–4 days were head-fixed to a custom mount with eicosane (Sigma) and imaged on Movable Objective Microscopes with galvanometric or resonant scanners (Sutter Instruments) controlled through ScanImage v.5.4.0 software (Vidrio Technologies). Cuticle, adipose tissue and trachea were removed to create an optical window, and the brain was superfused with carbogenated extracellular solution (95% O2–5% CO2, pH 7.3, 275 mOsm) containing 103 mM NaCl, 3 mM KCl, 5 mM TES, 8 mM trehalose, 10 mM glucose, 7 mM sucrose, 26 mM NaHCO3, 1 mM NaH2PO4, 1.5 mM CaCl2 and 4 mM MgCl2. To excite iATPSnFR and co-expressed tdTomato, Mai Tai DeepSee (Spectra Physics model eHP DS) or Coherent Chameleon Ultra II Ti:sapphire lasers produced 930-nm excitation light pulses whose power was modulated by Pockels cells (302RM, Conoptics). Emitted photons were collected by ×20 (1.0 numerical aperture (NA)) water immersion objectives (W-Plan-Apochromat, Zeiss), split into green and red channels by dichromatic mirrors (Chroma 565dcxr or Semrock BrightLine 565 nm) and detected by GaAsP photomultiplier tubes (H10770PA-40 SEL, Hamamatsu Photonics). The emission paths contained bandpass filters for iATPSnFR (Semrock BrightLine FF01-520/60) and tdTomato (Chroma ET605/70 m), respectively. Photocurrents were passed through high-speed amplifiers (HCA-4M-500K-C, Laser Components) and integrators (BLP-21.4+, Mini-Circuits, or EF506, Thorlabs) to maximize the signal-to-noise ratio.
To gate open the conductance of CsChrimson, a 625-nm LED (M625L3, ThorLabs) controlled by a dimmable LED driver (ThorLabs) delivered 0.5–25 mW cm−2 of optical power through a bandpass filter (Semrock BrightLine FF01-647/57-25) to the head of the fly. Stimulus trains lasted for 2 min and consisted of ten 25-ms light pulses in 500-ms bursts recurring once per s. To apply arousing heat, an 808-nm laser diode (Thorlabs L808P500MM) was mounted on a temperature-controlled heat sink (ThorLabs TCDLM9 with ThorLabs TED200C controller) and aimed at the abdomen of the fly. The diode was restricted to a maximal output of 50 mW by a ThorLabs LDC210C laser diode controller, and 2-s pulses were delivered every 30 s for 2 min. Images of 256 × 256 pixels were acquired at a rate of 29.13 Hz. The voltage steps controlling the LED or laser diode were recorded in a separate imaging channel for post-hoc alignment.
To drive proton pumping by mito-dR, a 530-nm LED (M530L3, ThorLabs) controlled by a dimmable LED driver (ThorLabs) delivered about 25 mW cm−2 of optical power through an ACP2520-A collimating lens (ThorLabs) to the head of the fly. Stimuli lasted for 400 ms and were repeated every 10 s. Two shutters, one in the combined fluorescence emission path (∅1/2″ stainless steel diaphragm optical beam shutter with controller, Thorlabs) and one on the LED (Vincent/UniBlitz VS35S2ZM1R1-21 Uni-Stable Shutter with UniBlitz VMM-T1 Shutter Driver/Timer controller), opened alternately during imaging and green light stimulation. Images of 128 × 50 pixels were acquired at a rate of 41.59 Hz. The voltage steps controlling the LED were recorded in a separate imaging channel for post-hoc alignment.
Time series of average fluorescence in manually selected dendritic regions of interest were analysed in MATLAB, following the subtraction of a time-varying background. ΔF/F curves were calculated separately for each trial as \(\frac{\Delta {F}_{t}}{{F}_{0}}=\frac{({F}_{t}-{F}_{0})}{{F}_{0}}\), where F0 is the mean fluorescence intensity before stimulation onset (170 s for heat and CsChrimson, 4 s for mito-dR) and Ft is the fluorescence intensity in frame t; \(\frac{\Delta {R}_{t}}{{R}_{0}}\) represents the iATPSnFR–tdTomato (green–red) intensity ratio. The stimulus-aligned \(\frac{\Delta {R}_{t}}{{R}_{0}}\) signals were averaged across two trials in the case of CsChrimson and 30 trials in the case of mito-dR and then across flies; the statistical units are flies. For display purposes, traces were smoothed with moving-average filters (15-s windows for heat and CsChrimson, followed by downsampling by a factor of 100; 1-s window for mito-dR).
Super-resolution and confocal imaging
Single-housed females aged 6 days post eclosion were dissected at zeitgeber time 0, following ad libitum sleep or 12 h of sleep deprivation. Experimental and control samples were processed in parallel. Brains were fixed for 20 min in 0.3% (v/v) Triton X-100 in PBS (PBST) with 4% (w/v) paraformaldehyde, washed five times with PBST, incubated with primary antibodies and secondary detection reagents where indicated, mounted in Vectashield and imaged. Only anatomically intact specimens from live flies (at the point of dissection) were analysed, blind to sleep history, using existing, adapted or newly developed (semi-)automated routines in Fiji 2.14.0/1.54f. The specific acquisition and analysis parameters for different experiments are as follows and further detailed in Supplementary Table 4.
For mitochondrial morphometry106, z-stacks of the dendritic fields of mito-GFP-expressing dFBNs (or the glomerular arborizations of projection neurons) were collected at the Nyquist limit, with identical image acquisition settings across all conditions. Dendrites were chosen as an optically favourable compartment for analysis because their branches (and the mitochondria within them) were well separated along all axes.
OPRM super-resolution images were collected on an Olympus IX83 P2ZF microscope (Olympus cellSense Dimension 4.3.1) equipped with a UplanSApo ×60 (1.30 NA) silicon oil objective and a Yokogawa CSU-W1 super-resolution by optical pixel reassignment spinning-disc module38. Image stacks were passed through a ‘low’ Olympus Super Resolution spatial frequency filter and deconvolved in five iterations of a constrained maximum likelihood algorithm (Olympus cellSense Dimension 4.3.1).
CLSM images were acquired on a Leica TCS SP5 confocal microscope (Leica LAS AF 2.7.3.9723) with an HCX IRAPO L ×25 (0.95 NA) water immersion objective. Point-spread functions were created with the PSF Generator plugin107 in Fiji and used to deconvolve the images with DeconvolutionLab2 2.1.2 software108,109 using the Richardson–Lucy algorithm with total variation regularization (set to 0.0001) and a maximum of two iterations106.
Functions of the Mitochondria Analyzer 2.3.1 plugin in Fiji were applied in an automated fashion to the deconvolved OPRM and CLSM images to remove background noise (‘subtract background’ with a radius of 1.25 μm), reduce noise and smooth objects while preserving edges (‘sigma filter plus’), enhance dim areas while minimizing noise amplification (‘enhance local contrast’ with slope 1.4) and optimize the use of image bits (‘gamma correction’ with a value of 0.90) before thresholding (‘weighted mean’ with block size 1.25 μm and a ‘C value’ of 5 for dFBNs and 12 for projection neurons, determined empirically to minimize background noise106). The resulting binary images were examined by means of Batch 3D analysis on a ‘per-cell’ basis to extract morphological metrics; OPRM image stacks were rendered in three dimensions with the Volume Viewer 2.02 plugin in Fiji, using trilinear interpolation in volume mode with default settings and a greyscale transfer function. The full dendritic fields of dFBNs were analysed, but the high packing density of projection neuron dendrites in the glomeruli of the antennal lobe forced us to select 20 substacks per glomerulus, each with an axial depth of 6 µm (for OPRM) or 5.8 µm (for CLSM), using a Fiji random number generator function, to make computations practical. Morphometric parameters were then averaged across all substacks per glomerulus.
When other UAS-driven transgenes were present in addition to UAS-mito-GFP, the dilution of a limiting amount of GAL4 among several promoters led to a noticeable dimming of mitochondrial fluorescence, especially in OPRM images. We therefore sought to compare mitochondrial morphologies within (Fig. 3a,b and Extended Data Figs. 5g and 6a,b) or among (Extended Data Figs. 5c,d and 7a,b) genotypes carrying the same number of UAS-driven transgenes. Although shape metrics, such as the mean volume, sphericity and branch length of mitochondria, seemed robust under variations in brightness, the number of fluorescent objects crossing the detection threshold was not. Because the absolute number of mitochondria in an image stack also depends on the volume fraction occupied by dFBN dendrites, these limitations should be borne in mind when interpreting mitochondrial counts.
For ratiometric snapshot imaging of the ATP sensors iATPSnFR (normalized to co-expressed RFP) and ATeam and the mitophagy sensor mito-QC, fluorescence was quantified on summed z-stacks of the relevant channels, following the subtraction of average background in manually defined areas close to the structures of interest. Brains expressing ATeam were imaged on a Zeiss LSM980 with Airyscan2 microscope (ZEN blue 3.3) with a Plan-Apochromat ×40 (1.30 NA) oil immersion objective, an excitation wavelength of 445 nm for the FRET donor mCFP, and emission bands of 454–507 nm for CFP and 516–693 nm for the YFP variant mVenus75. In ATP calibration experiments (see below), the biocytin-filled dFBN soma was identified after staining with Alexa Fluor 633 streptavidin (ThermoFisher, 1:600 in 0.3% PBST at 4 °C overnight) at excitation and emission wavelengths of 639 nm and 649–693 nm, respectively. Images of iATPSnFR plus RFP and mito-QC were acquired on a Leica TCS SP5 confocal microscope with HCX IRAPO L ×25 (0.95 NA) water and HCX PL APO ×40 (1.30 NA) oil immersion objectives, respectively. The excitation and emission wavelengths were 488 nm and 498–544 nm for iATPSnFR, 555 nm and 565–663 nm for RFP, and 488 nm and 503–538 nm, and 587 nm and 605–650 nm, for the GFP and mCherry moieties of mito-QC, respectively.
To convert the emission ratios of ATeam1.03NL into approximate ATP concentrations, we constructed a calibration curve in vivo by including 0.15, 1.5 or 4 mM MgATP, along with 10 mM biocytin, in the intracellular solution during whole-cell patch-clamp recordings. After dialysing the recorded dFBN for 30 min, the patch electrode was withdrawn, and the brain was recovered, fixed and processed for detection and ratiometric imaging of the biocytin-labelled soma. A standard curve was obtained by fitting the Hill equation, with a dissociation constant75 of 1.75 mM and a Hill coefficient74 of 2.1, to the YFP–CFP fluorescence ratios of dFBN somata containing known ATP concentrations.
SPLICS puncta82,110 were imaged on a Leica TCS SP5 confocal microscope with an HCX PL APO ×40 (1.30 NA) oil immersion objective, excitation and emission wavelengths of 488 and 500–540 nm, respectively, and analysed in Fiji with a custom macro based on the Quantification 1 and 2 plugins110. Only puncta exceeding 10 voxels were counted.
For localizing Drp1, we labelled the mitochondria of dFBNs with mito-GFP in Drp1::FLAG-FlAsH-HA flies94, whose genomic Drp1 coding sequence is fused in frame with a Flag-FlAsH-HA tag. Fixed brains were incubated sequentially at 4 °C in blocking solution (10% goat serum in 0.3% PBST) overnight, with mouse monoclonal anti-Flag antibody (anti-DDK; 1:1,000, OriGene) in blocking solution for 2–3 days, and with goat anti-Mouse Alexa Fluor 633 (1:500, ThermoFisher) in blocking solution for two days. The samples were washed five times with PBST before and after the addition of secondary antibodies, mounted and imaged on a Leica TCS SP5 confocal microscope with an HCX PL APO ×40 (1.30 NA) oil immersion objective. The excitation and emission wavelengths were 488 and 500–540 nm for mito-GFP and 631 and 642–690 nm for Alexa Fluor 633, respectively. dFBN somata were identified manually in the green channel, which was then thresholded, despeckled and binarized by an automated custom macro in Fiji. The mitochondria-associated fraction of endogenous Flag-tagged Drp1 in dFBNs was quantified in summed z-stacks as the proportion of red-fluorescent pixels in the somatic volume that colocalized with mitochondrial objects.
Electrophysiology
For whole-cell patch-clamp recordings in vivo, female flies aged 2–4 days post eclosion were prepared as for functional imaging, but the perineural sheath was also removed for electrode access. The GFP-labelled somata of dFBNs were visually targeted with borosilicate glass electrodes (8–10 MΩ) filled with internal solution (pH 7.3, 265 mOsm) containing: 10 mM HEPES, 140 mM potassium aspartate, 1 mM KCl, 4 mM MgATP, 0.5 mM Na3GTP, 1 mM EGTA and 10 mM biocytin. Signals were acquired at room temperature (23 °C) in current-clamp mode with a MultiClamp 700B amplifier (Molecular Devices), lowpass-filtered at 5 kHz, and sampled at 10 kHz using an Axon Digidata 1550B digitizer controlled through pCLAMP 11.2 (Molecular Devices). Series resistances were monitored but not compensated. Data were analysed using v.3.0c of the NeuroMatic package111 (http://neuromatic.thinkrandom.com) in Igor Pro 8.04 (WaveMetrics). Current–spike frequency functions were determined from voltage responses to a series of current steps (5-pA increments from −20 to 105 pA, 1 s duration) from a pre-pulse potential of −60 ± 5 mV. Spikes were detected by finding minima in the second derivative of the membrane potential trace. Spike frequencies were normalized to membrane resistances, which were calculated from linear fits of the steady-state voltage changes elicited by hyperpolarizing current steps. Only dFBNs firing more than one action potential in response to depolarizing current injections, with resting potentials below −30 mV and series resistances remaining below 50 MΩ throughout the recording, were characterized further. Spike bursts were defined as sets of spikes with an average intra-burst inter-spike interval (ISI) less than 50 ms and an inter-burst ISI more than 100 ms. These ISI thresholds were set after visual inspection of the voltage traces of all recorded neurons. Cells were scored as bursting if they generated at least one action potential burst during the series of depolarizing current steps.
Analysis of mitochondria in volume electron micrographs
The hemibrain v.1.2.1 connectome39 was accessed in neuPrint+ (ref. 112). The volumes of mitochondria in all R23E10-GAL4-labelled dFBNs113 and uniglomerular olfactory projection neurons39 in the left hemisphere (which contains mostly the dendritic compartments of projection neurons) were retrieved through Neo4j Cypher and neuPrint-python queries and analysed with custom MATLAB scripts. Supplementary Table 5 lists the identification numbers (Body_IDs) of all analysed cells.
Quantification and statistical analysis
With the exception of sleep measurements, no statistical methods were used to predetermine sample sizes. Flies of the indicated genotype, sex and age were selected randomly for analysis and assigned randomly to treatment groups if treatments were applied (for example, sleep deprivation). The investigators were blind to sleep history and/or genotype in imaging experiments but not otherwise. All behavioural experiments were run at least three times, on different days and with different batches of flies. The figures show pooled data from all replicates.
Gene expression levels were compared by Bonferroni-corrected two-sided Wilcoxon rank-sum test. Gene ontology enrichment was quantified using Fisher’s exact test in PANTHER (with a false discovery rate-adjusted significance level of P < 0.05) or ViSEAGO (with an unadjusted significance level of P < 0.001).
Behavioural, imaging and electrophysiological data were analysed in Prism v.10 (GraphPad) and SPSS Statistics 29 (IBM). All null hypothesis tests were two-sided. To control type I errors, P values were adjusted to achieve a joint α of 0.05 at each level in a hypothesis hierarchy; multiplicity-adjusted P values are reported in cases of several comparisons at one level. Group means were compared by t-test, one-way ANOVA, two-way repeated-measures ANOVA or mixed-effects models, as stated, followed by planned pairwise analyses with Holm–Šídák’s multiple comparisons test where indicated. Repeated-measures ANOVA and mixed-effect models used the Geisser–Greenhouse correction. Where the assumption of normality was violated (as indicated by D’Agostino–Pearson test), group means were compared by Mann–Whitney test or Kruskal–Wallis ANOVA, followed by planned pairwise analyses using Dunn’s multiple comparisons test, as indicated. Frequency distributions were analysed by χ²-test, and categories responsible for pairwise differences114 were detected by locating cells with standardized residuals ≥2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.