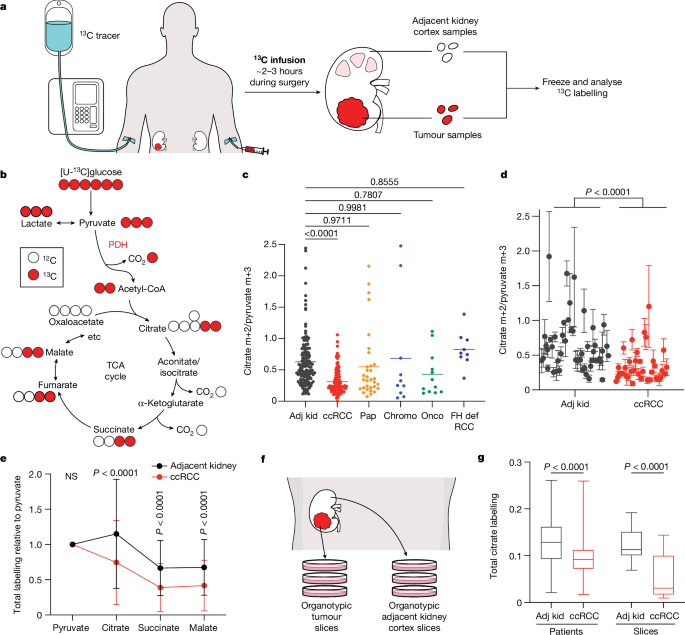
Patient infusions
The Institutional Review Board (IRB) at the University of Texas Southwestern Medical Center monitored and approved all conducted human participant research. Study protocols are subjected to annual continuing reviews by the IRB. All patients were recruited at the University of Texas Southwestern Medical Center. From December 2018 to August 2019, patients were enrolled on protocol STU062010-157. From September 2019 to present, patients are enrolled on protocol STU2019-1061. Two patients were enrolled on protocol STU052012-065. Generally, patients 18 years of age or older with radiographic evidence of known or probable kidney cancer requiring surgical biopsy or excision were recruited to an IRB-approved study and informed consent was obtained from all patients. Full eligibility and exclusion criteria, as well as recruitment procedures, are detailed in the Reporting Summary. Patients receiving [U-13C]glucose were enrolled on protocol STU062010-157 (NCT01668082), STU2019-1061 (NCT04623502) or STU052012-065 (NCT02095808) and infused at the following rate: 8âg bolus of [U-13C]glucose administered over 10âmin, followed by a continuous infusion of [U-13C]glucose at a rate of either 4 or 8âgâper hour. Patients receiving [1,2-13C]acetate were enrolled on protocol STU2019-1061 and were infused at the following rate: a bolus of 3âmg [1,2-13C]acetateâper kgâbody weight per minute for 5âmin, followed by a continuous infusion of [1,2-13C]acetate at 1.5âmgâper kg body weight perâminute. Patients receiving [U-13C]glutamine were enrolled on protocol STU2019-1061 and were infused at the following rate: a primer dose for 5âmin at a rate of 0.6âmg perâkg body weight perâminute, followed by a continuous infusion of 5.0âμmolâper kgâbody weight per minute (0.73âmgâper kg body weight perâminute). Uncontrolled or poorly controlled diabetes and pregnancy were exclusion criteria for the study. Demographic, clinical and pathological details are summarized in Supplementary Data Table 1. Once the specimens were removed from the body cavity, tissue samples were acquired with the attending pathologist or pathology assistant.
Animal studies
All procedures were approved by UT Southwestern Medical Centerâs Animal Care and Use Committee (protocol 2016-101360) or the University of Texas MD Anderson Cancer Centerâs Animal Care and Use Committee (protocol 00001158) in accordance with the Guide for the Care and Use of Laboratory Animals. Sample sizes were not pre-determined based on statistical power calculations but were based on our experience with these assays. All mice were given a numerical identifier, and tumour measurements were recorded according to the numerical identifier and unblinded at the end of the study. Mouse experiments requiring pharmacological treatments were not blinded to monitor mice for side effects. Mice were randomly allocated for injection and/or treatment.
Cell lines
786-O, Caki-1 and primary renal proximal tubule epithelial cells were purchased from the American Type Culture Collection (ATCC; CRL-1932, HTB-46 and PCS-400-010, respectively) and were authenticated by the ATCC. HEK293FT cells were purchased from Thermo Fisher (R70007) and were not authenticated after purchase. Metlow, Methigh, Methigh-50 and Methigh-26 were established from somatic mosaic genetically engineered mouse models (SM-GEMM) of kidney cancer36. Metlow and Methigh cell lines were established from Setd2KO;NF2KO;4q9p21â mice, and Methigh-50 and Methigh-26 cell lines were established from Setd2KO;VhlKO;4q9p21â mice. The Metlow cell line originated from a SM-GEMM mouse with no metastases, and the Methigh, Methigh-50 and Methigh-26 lines originated from SM-GEMM mice with ten or more metastases.
The primary kidney tumours were dissociated with a combination of mechanical dissociation and enzymatic dissociation using a 2âmgâmlâ1 mixture of collagenase IV/dispase (17104-019 and 17105-041, respectively, Invitrogen) resuspended in DMEM. Tissues were incubated for 1âh at 37â°C with trituration every 15âmin. Cells were then plated on plates coated with 0.1% gelatin and cultured in DMEM supplemented with 20% FBS and 1% penicillinâstreptomycin. To confirm metastatic potential, cells were reinjected under the renal capsule of NU-Foxn1nu mice as described in the âMetastatic colonization experiments using mouse cell lines in miceâ section, and mice were analysed for metastatic burden after 21 days. Primary tumours with high or low metastatic burden were dissociated and reinjected again under the renal capsule to select for differential metastatic capacity. Primary tumours that gave rise to a high number of metastatic nodules were termed Methigh, and clones that gave rise to fewer metastases were termed Metlow. The Methigh, Methigh-50, Methigh-26 and Metlow cell lines reflected the metastatic potential of their SM-GEMM of origin. Cells were kept in culture for five passages or less before re-implantations.
All cell lines were confirmed to be mycoplasma free using a commercial kit (2523348, Bulldog Bio). Cells were maintained in RPMI supplemented with 10% fetal bovine serum or 10% dialysed human serum and cultured at 37â°C in 5% CO2 and 95% air, unless otherwise noted.
High-resolution mass spectrometry (Q-TOF)
Data acquisition from isolated mitochondria and patient tissues for metabolomics was performed by reverse-phase chromatography on a 1290 UHPLC liquid chromatography system interfaced to a high-resolution mass spectrometry (HRMS) 6550 iFunnel Q-TOF mass spectrometer (Agilent Technologies). Frozen tissue fragments weighing 10â30âmg were added to ice-cold 80:20 methanol:water and extracted for metabolomics analysis. Samples were subjected to three freezeâthaw cycles, then centrifuged at 16,000g for 20âmin to precipitate macromolecules. The supernatant was evaporated using a vacuum concentrator. Samples were resuspended in 100âμl of 0.1% formic acid in water, vortexed for 30âs and centrifuged at 16,000g for 15âmin. Supernatant was transferred to an autosampler vial and then run on the mass spectrometer. The mass spectrometer was operated in both positive and negative (ESI+ and ESIâ) modes. Analytes were separated on an Acquity UPLC HSS T3 column (1.8âμm, 2.1âÃâ150âmm; Waters). The column was kept at room temperature. Mobile phase A composition was 0.1% formic acid in water, and mobile phase B composition was 0.1% formic acid in 100% acetonitrile. The liquid chromatography gradient was 1% phase B (5âmin), 5% phase B (10âmin), 99% phase B (8âmin), 99% phase B (1âmin), 1% phase B (1âmin) and 1% phase B (1âmin). The flow rate was 250âμlâminâ1. The sample injection volume was 5âμl.
ESI source conditions were set as follows: dry gas temperature of 225â°C and flow of 18âlâminâ1, fragmentor voltage of 175âV, sheath gas temperature of 350â°C and flow of 12âlâminâ1, nozzle voltage of 500âV and capillary voltage of +3,500âV in positive mode and â3,500âV in negative mode. The instrument was set to acquire over the full m/z range of 40â1,700 in both modes, with the mass spectrometer acquisition rate of 1 spectrum per second in profile format.
Raw data files (.d) were processed using Profinder B.08.00 SP3 software (Agilent Technologies) with an in-house database containing retention time and accurate mass information from 600 standards from Mass Spectrometry Metabolite Library (IROA Technologies). The database was created under the same data acquisition conditions described above. The in-house database matching parameters were: mass tolerance of 10âppm and retention time tolerance of 0.5âmin. Peak integration results were manually reviewed in Profinder and exported as a spreadsheet (.csv).
Metabolomics analysis
The signal-to-noise ratio for each metabolite was calculated by dividing the median of the quality control samples by the median of the blank samples. Following this, a run order correlation test was performed to determine the correlation between the run order and metabolite intensity for quality control samples injected at fixed intervals, adjusting for multiple comparisons using the BenjaminiâHochberg procedure. A LOESS (locally estimated scatterplot smoothing) correction was then applied to adjust for trends in the data that correlate with run order, which involved fitting a LOESS model to the quality control data and adjusting the sample data accordingly only if the trend in quality control samples within the interquartile range was statistically significant. Normalization was then performed using total ion count, using a subset of stable metabolites with low coefficient of variation and high signal-to-noise ratio to calculate a normalization factor based on their median intensities. This was used to adjust the intensity values of all metabolites in a sample. After normalization, a log10 transformation was applied to obtain a normal distribution. In cases in which a metabolite was measured in both positive and negative modes, the measurement with the lower signal-to-noise ratio was removed.
We compared our metabolomics dataset to a previously reported ccRCC dataset14. The value of each metabolite was normalized to the minimum observed value across the dataset. A log10 transformation was applied to the normalized values to stabilize variance and render the data more suitable for parametric analysis. Cohenâs d was used to assess the differential abundance of metabolites between tumour and adjacent normal tissues. For integration and comparison with the Hakimi dataset, we standardized the metabolite nomenclature by mapping the metabolite names to their respective Human Metabolome Database (HMDB) identifiers50. For the 152 metabolites that were common between both studies, the Pearson correlation coefficient of the effect sizes was calculated to assess the concordance of datasets.
Raw metabolomics files are available at the US National Institutes of Health Common Fundâs National Metabolomics Data Repository website, the Metabolomics Workbench51 (https://www.metabolomicsworkbench.org/), where it has been assigned project ID PR001954. A direct link to the data can be found in the âData availabilityâ section.
HRMS (Orbitrap)
[1,2-13C]acetate-infused tissue samples were analysed using an Orbitrap Fusion Lumos 1M Tribrid Mass Spectrometer. HILIC chromatographic separation of metabolites was achieved using a Millipore ZIC-pHILIC column (5âμm, 2.1âÃâ150âmm) with a binary solvent system of 10âmM ammonium acetate in water, pH 9.8 (solvent A), and acetonitrile (solvent B) with a constant flow rate of 0.25âmlâminâ1. For gradient separation, the column was equilibrated with 90% solvent B. After injection, the gradient proceeded as follows: 0â15âmin of linear ramp from 90% B to 30% B; 15â18âmin of isocratic flow of 30% B; 18â19âmin of linear ramp from 30% B to 90% B; 19â27 column regeneration with isocratic flow of 90% B. HRMS data were acquired with two separate acquisition methods. Individual samples were acquired with an HRMS full scan (precursor ion only) method, switching between positive and negative polarities. For data-dependent, high-resolution tandem mass spectrometry methods, precursor ion scans were acquired at a resolving power of 120,000 full width at half-maximum (FWHM) with a mass range of either 50â750 or 70â1,050âDa. The AGC target value was set to 1âÃâ106 with a maximum injection time of 100âms. Pooled samples were generated from an equal mixture of all individual samples and analysed using individual positive-polarity and negative-polarity spectrometry data-dependent, high-resolution tandem mass spectrometry acquisition methods for high-confidence metabolite ID. Product ion spectra were acquired at a resolving power of 15,000 FWHM without a fixed mass range. The AGC target value was set to 2âÃâ105 with a maximum injection time of 150âms. Data-dependent parameters were set to acquire the top ten ions with a dynamic exclusion of 30âs and a mass tolerance of 5âppm. Isotope exclusion was turned on, and a normalized collision energy value of 30% was used or a stepped normalized collision energy applied with values of 30%, 50% and 70%. Settings remained the same in both polarities. Metabolite identities were confirmed in three ways: (1) precursor ion m/z was matched within 5âppm of theoretical mass predicted by the chemical formula; (2) fragment ion spectra were matched within a 5âppm tolerance to known metabolite fragments; and (3) the retention time of metabolites was within 5% of the retention time of a purified standard run with the same chromatographic method. Metabolites were relatively quantitated by integrating the chromatographic peak area of the precursor ion searched within a 5âppm tolerance.
Acetyl-CoA fractional enrichment was determined with a selected ion monitoring (SIM) scan event on an Orbitrap Fusion Lumos 1M Tribrid Mass Spectrometer. The SIM scan event targeted the theoretical mass for the positive ion of acetyl-CoA in positive ionization mode (m/z 810.1330) with a 4.5-Da window. Data were collected with a resolving power of 60,000 FWHM with an AGC target of 4âÃâ105 ions. To calculate fractional enrichment of m+2 acetyl-CoA, the SIM scan integrated the m+0, m+1 and m+2 peaks and the full-scan data to integrate the remaining naturally abundant isotopes. Isotope enrichment was corrected for natural abundance.
Mass spectrometry for isotopomer analysis
Samples were analysed on an AB Sciex 6500 QTRAP liquid chromatography/mass spectrometer (Applied Biosystems SCIEX) equipped with a vacuum degasser, quaternary pump, autosampler, thermostatted column compartment and triple quadrupole/ion trap mass spectrometer with electrospray ionization interface, and controlled by AB Sciex Analyst 1.6.1 Software. SeQuant ZIC-pHILIC 5-µm polymer (150âÃâ2.1âmm) columns were used for separation. Solvents for the mobile phase were 10âmM ammonium acetate aqueous (pH 9.8 adjusted with NH3â¢H2O (A) and pure acetonitrile (B). The gradient elution was: 0â20âmin, linear gradient of 90â65% B; 20â23âmin, linear gradient of 65â30% B; 23â28âmin, 30% B; and 28â30âmin, linear gradient of 30â90% B, then reconditioning the column with 90% B for 5âmin. The flow rate was 0.2âmlâminâ1 and the column was operated at 40â°C. Glutamate isotopomers were analysed using a published method17.
Gas chromatographyâmass spectrometry
Gas chromatographyâmass spectrometry was used to analyse infused patient tissue and plasma samples, as well as tracing assays in cell lines and slice cultures. Blood was obtained before and approximately every 30âmin during infusion, when congruent with surgical workflow, until tissue was removed from the patient. Whole blood was chilled on ice and centrifuged (at 1,500g for 15âmin at 4â°C, with acceleration and deceleration rates set to 5) to separate and freeze the plasma. Aliquots (50âµl) of plasma were added to ice-cold 80:20 methanol:water for extraction. Frozen tissue fragments weighing roughly 10â30âmg were added to ice-cold 80:20 methanol:water and extracted to analyse 13C enrichment. Samples were subjected to three freezeâthaw cycles, then centrifuged at 16,000g for 20âmin to precipitate macromolecules. The supernatant was evaporated using a vacuum concentrator and resuspended in 30âμl of methoxyamine (10âmgâmlâ1) in pyridine. Samples were transferred to autoinjector vials and heated at 70â°C for 15âmin. A total of 70âμl of tert-butyldimethylsilyl was added, and the samples were briefly vortexed and heated for another 60âmin at 70â°C. Injections of 1âμl were analysed on an Agilent 7890A gas chromatograph coupled to an Agilent 5975C mass selective detector. The observed distributions of mass isotopologues were corrected for natural abundance52.
mtDNA:nuclear DNA quantitative PCR
Genomic DNA was isolated using the DNeasy Blood & Tissue Kit (69504, Qiagen). Samples were run using the Luna Universal One-Step qPCR Kit (M3003, New England Biolabs) on a CFX384 machine (Bio-Rad). The following primers were used for human COX2 as representative of mtDNA: CCGTCTGAACTATCCTGCCC (forward), (GCCGTAGTCGGTGTACTCGT (reverse). The following primers were used for human histone 3 (H4C3) as representative of nuclear DNA: GGGATAACATCCAGGGCATT (forward), CCCTGACGTTTTAGGGCATA (reverse).
RNA isolation
RNA was isolated using TRIzol (15596018, Thermo Fisher) and an RNeasy Mini Kit (74106, Qiagen). Total RNA was quantified using a Qubit fluorometer (Invitrogen) and the Invitrogen Qubit RNA High Sensitivity kit (Q32852, Invitrogen). Samples were diluted in ultrapure water before sequencing.
NDI1 RTâquantitative PCR
RNA was isolated as described above and samples were run using the Luna Universal One-Step RTâqPCR Kit (E3005, New England Biolabs) on a CFX384 machine (Bio-Rad). The following primers were used for NDI1: GCCGAAGAAGTCCAAATTCAC (forward), CGACAGCCGTTCTCAGAT (reverse). The following primers were used for ACTB (encoding β-actin): CTAAGGCCAACCGTGAAAAG (forward), ACCAGAGGCATACAGGGACA (reverse).
RNA sequencing
RNA sequencing libraries were prepared using the NEBNext Ultra II directional RNA library prep kit with the NEBNext poly(A) mRNA magnetic isolation module (E7490L and E7760L, New England Biolabs) according to manufacturerâs instructions. Libraries were stranded using standard NEB indices according to the manufacturerâs instructions (E7730L, E7335L and E7500L, New England Biolabs). Sequencing reads were aligned to the human reference genome (hg19) by STAR 2.7.3.a with default parameters in the two-pass mode. Counts for each gene were generated using htseq-count v0.6.1. Differentially expressed genes were identified by DESeq2 v1.14.1. The ends of sequences were trimmed with remaining adapter or quality scores of less than 25. Sequences less than 35âbp after trimming were removed. The trimmed Fastq files were aligned to GRCh38 using HiSAT2 (ref. 53) and duplicates were marked with SAMBAMBA. Features (genes, transcripts and exons) were counted using featureCounts54. Differential expression analysis was performed using EdgeR55 and DESeq56. Raw sequencing files are deposited on the Gene Expression Omnibus (GSE251905). Extended Data Fig. 1a compares RNA sequencing data from the TCGA cohort and this study, emphasizing genes related to the ETC and glycolysis. The Cohenâs effect size (d) between tumour and adjacent kidney for each of 15,642 genes was correlated between TCGA data and data from the current cohort. ETC genes were selected from the gene ontology cellular component library, including genes related to complexes IâIV of the ETC. The glycolysis genes include the following four gene sets: KEGG_GLYCOLYSIS_GLUCONEOGENESIS, REACTOME_GLYCOLYSIS, HALLMARK_GLYCOLYSIS and WP_GLYCOLYSIS_AND_GLUCONEOGENESIS.
ESTIMATE analysis
The ESTIMATE R package57 was used to derive stromal and immune scores and a tumour purity ESTIMATE score from RNA sequencing data.
OxPhos score calculation
A list of OxPhos genes from the MSigDB database58 was created by combining the genes in the KEGG_OXIDATIVE_PHOSPHORYLATION gene set with the genes encoding subunits of the PDH complex (PDHA1, PDHB, DLAT, DLD and PDHX). Principal component analysis was performed on log2-transformed, mean-centred and z-transformed data, and the first principal component (PC1) was extracted and used as the OxPhos score.
Organotypic slice cultures
After surgery, kidney cortex and tumour fragments were embedded in 0.1% agarose (BP1356, Thermo Fisher) and sliced into approximately 300-μM thick sections using a microtome (Compresstome, VF-300, Precisionary Instruments). These tissues were then transferred and maintained on hydrophilic polytetrafluoroethylene cell culture inserts (PICM0RG50, Millipore) in human plasma-like medium supplemented with 10% dialysed human serum. Before tracing assays, tissues were washed twice with 0.9% saline, and medium was replaced with human plasma-like medium without unlabelled glucose and replaced with an equivalent concentration of [U-13C]glucose for 3âh. Slices were maintained in an incubator with 5% CO2, 5% O2 and 90% N2 at 37â°C.
Human serum dialysis
Human serum was purchased from Sigma-Aldrich (H3667) and dialysed using SnakeSkin dialysis tubing, 3.5âK MWCO, 35âmm (PI88244, Thermo Fisher). Serum was dialysed against a 20X volume of PBS. Dialysis was performed for 48âh at 4â°C with a complete PBS exchange every 9â12âh. Dialysed serum was then sterile filtered using bottle-top vacuum filters with a pore size of 0.22âμm (431097, Corning).
Mitochondrial isolation and respiration measurements
OCRs were measured using a Seahorse Xfe96 Analyzer (Agilent Technologies) as previously described59,60. Fresh kidney and tumour samples were homogenized with 40 strokes of a Dounce homogenizer in mitochondrial isolation buffer consisting of 5âmM HEPES, 70âmM sucrose, 220âmM mannitol, 5âmM MgCl2, 10âmM KH2PO4 and 1âmM ethylene glycol-bis(β-aminoethyl ether)-N,N,Nâ²,Nâ²-tetraacetic acid (EGTA), pH 7.2; H4034, S0389, M4125, 208337, P9541 and E3889, respectively, Sigma-Aldrich) and isolated via differential centrifugation at 4â°C. Nuclei and cell debris were removed by centrifuging five times at 600g. Mitochondria were pelleted with a 10,000g spin and washed twice. Mitochondria were quantified using a detergent compatible assay (5000112, Bio-Rad).
Outer mitochondrial membrane fidelity was tested using a proteinase K assay as previously described61. In brief, 50âμg mitochondria were incubated on ice for 10âmin with proteinase K (AM2542, Thermo Fisher) at a final concentration of 10âμgâmlâ1. The reaction was stopped by pelleting mitochondria and resuspending in lysis buffer (2% SDS and 25âmM Tris. pH 6.8) with 1âmM phenylmethylsulfonyl fluoride (PMSF; 36978, Thermo Fisher) and immediately snap frozen in liquid nitrogen. Immunoblots were then run on the proteinase K-treated mitochondria as described in the âImmunoblottingâ section below for both an outer mitochondrial membrane protein (anti-Tom20, 1:2,000 dilution; 11802, Proteintech) and a mitochondrial matrix protein (anti-HSP60, 1:2,000 dilution; 12165S, Cell Signaling Technology). For experiments reported in this paper, we only used respiration and tracing data from mitochondria with an intact outer mitochondrial membrane (as evidenced by an intact, non-smeared HSP60 band after proteinase K treatment).
Of mitochondria, 5âµg was plated in an Xfe96 plate on ice and centrifuged at 2,700g for 2âmin at 4â°C. Isolation buffer containing ETC complex substrates was added to cells and measurements were started immediately. The following ETC complex substrates were used: complex Iâpyr/mal (10âmM pyruvate and 1âmM malate), complex Iâglu/mal (10âmM glutamate and 1âmM malate), complex II (5âmM succinate and 4 μM rotenone) and complex IV (10âmM ascorbate, 100 μM TMPD and antimycin A 2âμM). At indicated times, 4âmM ADP, 2âμM oligomycin A, 2âμM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) and either 4âμM antimycin A or 40âμM sodium azide were injected. All chemicals were purchased from Sigma-Aldrich (P2256 (pyruvate), 240176 (malate), 49621 (glutamate), S3674 (succinate), R8875 (rotenone), A4403 (ascorbate), T7394 (TMPD), A8674 (antimycin A), A5285 (adenosine diphosphate), 75351 (oligomycin A), C2759 (CCCP) and S2002 (sodium azide)). All indicated concentrations are final concentrations. Respiratory control ratios were calculated by state III/state IV respiration.
For cells expressing the pMXs-EV or pMXs-NDI1 retroviral vector, cells were plated in a 96-well plate at a concentration of 2âÃâ104 cells per well in 80âμl RPMI-1640 media with 4âmM glutamine and 10% FBS. Cells were incubated in a CO2-free incubator at 37â°C for 1âh before Xfe96 measurements to allow temperature and pH equilibration. Seahorse assays consisted of three mix (3âmin) and measurement (3âmin) cycles, allowing determination of OCR and the extracellular acidification rate every 6âmin.
NDI1, LbNOX and dsRed luciferase expression
pMXs-EV, pMXs-mito-LbNOX and pMXs-cyto-LbNox were a gift from J. Garcia-Bermudez62. pMXs-NDI1 was a gift from D. Sabatini (plasmid 72876, AddGene)63. pMXs-EV, pMXs-NDI1, pMXs-mito-LbNOX or pMXs-cyto-LbNOX was transfected into 293FT cells with gag-pol and VSVG using Lipofectamine 3000 (L3000015, Thermo Fisher) or PolyJet (SL100688, SignaGen) according to the manufacturerâs instructions. Viral supernatants were collected 48âh after transfection and filtered through a 0.45-μm filter. Cells were cultured with virus containing media and 4âµgâmlâ1 polybrene (TR-1003-G, Sigma-Aldrich) for 24âh, after which fresh medium was added. Cells were then exposed to 10âµgâmlâ1 blasticidin selection until uninfected cells died.
A bi-cistronic lentiviral construct carrying dsRed2 and luciferase (dsRed2-P2A-Luc) was a gift from S.J. Morrison. dsRed2-P2A-Luc with pMD2G and psPAX2 were transfected into 293FT cells using Polyjet (SL100688, SignaGen) according to the manufacturerâs instructions. Viral supernatants were collected 48âh after transfection and filtered through a 0.45-μm filter. Cells with either pMXs-EV, pMXs-NDI1, pMXs-cyto-LbNOX or pMXs-mito-LbNOX were cultured with virus containing media and 4âµgâmlâ1 polybrene (TR1003, Sigma-Aldrich) for 8âh, after which the medium was changed to fresh medium. The top 10% of live cells expressing dsRed were sorted using fluorescence-activated cell sorting (FACS). 786-O and Caki-1 cells expressing dsRed were trypsinized and filtered through a 40-μm cell strainer to obtain a single-cell suspension and stained with 4â²,6-diamidino-2-phenylindole (DAPI; 62248, Thermo Fisher). Cells were gated to exclude dead cells, cell debris and doublets based on FSC/SSC, then gated on live cells (DAPI-negative cells). The top 10% of dsRed-positive cells were sorted, collected and used for experiments. Example FACS plots exemplifying the gating strategy are provided in Supplementary Fig. 2. Data were analysed using BD FACSDiva 8.0 and FlowJo V10.
Immunofluorescence and confocal microscopy
Coverslips were coated with 10âµgâmlâ1 fibronectin (F1141, Sigma-Aldrich) for 1âh at 37â°C and rinsed once with PBS. Cells were immediately seeded on the coverslips and fixed the next day with fresh warm 4% paraformaldehyde solution in PBS for 15âmin followed by permeabilization using 0.1% (v/v) Triton X-100 in PBS at room temperature for 10âmin. Cells were blocked in filtered PBS containing 1% BSA for at least 30âmin at room temperature before incubation with primary antibodies to FLAG (1:200 dilution; F1804, Sigma-Aldrich) and HSP60 (1:500 dilution; 12165S, Cell Signaling Technology) for 1âh at room temperature. Cells were washed three times for 5âmin with PBS and incubated with fluorophore conjugated secondary antibodies (1:500 dilutions; 111-545-144, Jackson ImmunoResearch Laboratories and A31570, Thermo Fisher) for 1âh at room temperature in the dark. Coverslips were washed with PBS three times for 5âmin and Mili-Q water once before being mounted on Profade-Antifade (P36935, Invitrogen) slides overnight in the dark. Cells were imagined using a Zeiss LSM 880 confocal laser scanning microscope with Z-stacks acquired. All representative images were processed using ImageJ.
Ki67 staining
After removal of tumour-bearing organs, tissues were fixed in 10% formalin, sectioned and stained for Ki67-positive nuclei (1:500 dilution; MA5-14520, Thermo Fisher). Slides were imaged using an inverted Zeiss LSM780 confocal microscope. Ki67-positive nuclei were quantified using ImageJ.
Immunoblotting
Cells were lysed in RIPA buffer (BP-115, Boston BioProducts) containing protease and phosphatase inhibitors (78444, Thermo Fisher), then centrifuged at 4â°C for 10âmin at approximately 20,160g. Supernatants were transferred to new pre-chilled 1.5-ml tubes, and protein concentrations were quantified using a BCA Assay Kit (23225, Thermo Fisher). Protein lysates were resolved via SDSâPAGE and transferred to PVDF membranes. Membranes were blocked in 5% BSA in Tris buffered saline with Tween-20 (TBST; 20âmM Tris (pH 7.5), 150âmM NaCl and 0.1% Tween-20) and then incubated with primary antibody (anti-FLAG M2, 1:2,000 dilution; F1804, Sigma-Aldrich) in TBST supplemented with 5% BSA at 4â°C overnight. The primary antibody was detected with a horseradish peroxidase-conjugated secondary antibody (1:2,000 dilution; 7076S, Cell Signaling Technology) for 1âh followed by exposure to ECL reagents (PI32106, Fisher Scientific). After imaging, membranes were stripped with Restore Striping Buffer (21059, Thermo Fisher), washed with TBST three times and blocked with 5% BSA in TBST for 1âh. Membranes were then incubated with primary antibody (β-actin, 1:2,000 dilution; 8457S, Cell Signaling Technology) in TBST supplemented with 5% BSA at 4â°C overnight. The primary antibody was detected with a horseradish peroxidase-conjugated secondary antibody (1:2,000 dilution; 7074S, Cell Signaling Technology) for 1âh followed by exposure to ECL reagents (PI32106, Thermo Fisher).
[U-13C]glucose tracing in cell lines
[U-13C]glucose tracing data from non-small-cell lung cancer cell lines were previously reported41. Similar assay conditions described below were used for tracing experiments in this study. Before tracing experiments, cells expressing either empty vector or NDI1 were washed twice with 0.9% saline, and medium was replaced with RPMI-1640 containing [U-13C]glucose supplemented with 5% dialysed FBS for 6âh. Cells were rinsed in ice-cold 0.9% saline and lysed with three freezeâthaw cycles in ice-cold 80% methanol. Samples were then prepared for gas chromatographyâmass spectrometry analysis.
Subcutaneous implantation of human cell lines in mice
Cell suspensions were prepared for injection in a 1:1 mixture of Matrigel (354234, Corning) to staining medium (Leibovitzâs L15 medium (21083027, Thermo Fisher), 1âmgâmlâ1 BSA (A2153, Sigma-Aldrich), 1% penicillinâstreptomycin (P0781, Sigma-Aldrich) and 10âmM HEPES (pH 7.4)). Subcutaneous injections were performed in NOD.CB17-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice in a final volume of 100âμl. Four-to-eight-week-old male and female NSG mice were transplanted with 1,000,000 cells subcutaneously in the right flank. Both male and female mice were used. For all subcutaneous experiments, the maximum permitted tumour diameter was 2.0âcm, which was not exceeded in any experiment.
Metastatic colonization experiments using human cell lines in mice
Cell suspensions were prepared for injection in staining medium described above. Tail-vein injections were performed in NOD.CB17-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice in a final volume of 50âμl. Four-to-eight-week-old male and female NSG mice were transplanted with 250,000 cells. Both male and female mice were used. Metastatic burden was assessed weekly by bioluminescence. Five minutes before performing luminescence imaging, mice were injected intraperitoneally with 100âμl of PBS containing 40âmgâmlâ1 d-luciferin monopotassium salt (L8220, Biosynth) and mice were anaesthetized with isoflurane 2âmin before imaging. The mice were imaged using an IVIS Imaging System 200 Series (Caliper Life Sciences) or an Ami HTX (Spectral Instruments Imaging). The exposure time ranged from 10 to 60âs, depending on the maximum signal intensity, with the 60-s timepoint used for greater range only if the 30-s timepoint was not saturated. The bioluminescence signal (total photon flux) was quantified with âregion of interestâ measurement tools in Living Image software (Perkin Elmer) or Aura Imaging software (Spectral Instruments Imaging). The maximal tumour burden was defined as a bioluminescent signal saturated after 30âs, which was not exceeded during any experiment.
Metastatic colonization experiments using mouse cell lines in mice
Maximal tumour burden was not exceeded according to the IRB guidelines. For orthotopic tumours, mice were euthanized 21 days after implantation or after they developed symptoms of distress. Both male and female mice were used. Mice were kept in a 12-h lightâ12-h dark cycle as commonly used, and housed at 18â23â°C with humidity of 50â60%.
To establish orthotopic tumours, 10,000 cells were resuspended in a 2:1 solution of OPTI-MEM (31985062, Gibco) and Matrigel (354234, Corning). Six-to-nine-week-old NU-Foxn1nu (Jackson Laboratories) mice were anaesthetized using isoflurane. Buprenorphine slow release (0.1âmgâkgâ1 two times daily) was subcutaneously injected, and shaved skin was disinfected with 70% ethanol and betadine (1425, Dynarex). A 1-cm incision was performed on the left flank through the skin/subcutaneous and muscular/peritoneal layers. The left kidney was exposed and 20âμl of the cell suspension was injected under the kidney capsule. The kidney was repositioned into the abdominal cavity, and muscular/peritoneal planes were closed individually by absorbable sutures. The skin/subcutaneous planes were closed using metal clips. Mice were monitored daily for the entire duration of the experiment. Twenty-one days after orthotopic kidney implantation, mice were imaged for primary tumour burden in the kidney and metastatic tumour burden in the lungs. A 7T Bruker Biospec (BrukerBioSpin), equipped with a 35-mm inner-diameter volume coil and 12-cm inner-diameter gradients, was used for MRI. A fast acquisition with relaxation enhancement sequence with 2,000/39-ms TR/TE (repetition time/echo time), 256âÃâ192 matrix size, r156-µM resolution, 0.75-mm slice thickness, 0.25-mm slice gap, 40âÃâ30-cm2 field of view, 101-kHz bandwidth and 4 number of excitation was used to acquire multislice T2-weighted images in coronal and axial planes. At end point, mice were euthanized by exposure to carbon dioxide followed by cervical dislocation. A necropsy form was filled in with mouse information, tumour size, and metastasis location and number. Fluorescent and brightfield images were acquired through a Leica MZ12s stereo microscope. TdTomato-positive lesions were quantified using LAS v4.13 software (Leica Microsystems).
IACS-010759 treatment
Mice were allowed to recover from implantation surgery. The following day, mice were treated with either 100âμl of vehicle or IACS-010759 (S8731, Selleckchem) dissolved in vehicle via oral gavage on a 5-day on, 2-day off schedule. The vehicle was composed of 5% DMSO, 40% PEG300, 5% Tween-80, and 50% ddH2O. IACS-010759 was dosed at 5âmgâkgâ1. Mice were euthanized after 21 days in accordance with IRB guidelines.
CRISPR screening in Methigh and Metlow cells
Lentiviral particles of the mouse genome-wide CRISPR library (mTKOv3) were generated by the University of Michigan Biomedical Research Lentiviral Core and concentrated 100Ã. Cells were transduced with the mouse genome-wide CRISPR library in 500-cm2 square dishes with 8âμgâmlâ1 polybrene at a multiplicity of infection of 0.3 and an estimated 400à coverage. The medium was replaced 24âh after infection, and puromycin selection started at 48âh. At 72âh, cells were trypsinized, pooled and counted. As a reference, 30âÃâ106 cells were immediately collected. Every passage of 15âÃâ106 cells (approximately 200à coverage) was maintained in culture until the end point (20 doublings) when 30âÃâ106 cells (approximately 400à coverage) were collected.
Cell pellets were suspended in 2âml of buffer P1/RNAse A and lysed by adding 1/20 volume of 10% SDS. Genomic DNA was sheared by passing the lysate 10â15 times through a 22-gauge syringe needle after 10âmin. One volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added to the lysate, and the samples were centrifugated at 17,000g for 10âmin, and the upper phase was transferred to a new tube. The second extraction step was done with chloroform:isoamyl alcohol (24:1), and the upper phase was transferred to a new tube and mixed with 0.1 volumes of 3âM NaCl and 0.8 volumes of 2-propanol to precipitate genomic DNA. Samples were centrifuged at 17,000g for 20âmin at 4â°C, and the DNA pellet was washed in 70% ethanol and centrifuged for 5âmin at 17,000g at 4â°C. The DNA pellet was then dried and resuspended overnight in UltraPure distilled water. The genomic DNAs were quantified using a NanoDrop 2000 (Thermo Fisher). For the generation of next-generation sequencing libraries, barcodes were amplified in two rounds of PCR using the Titanium Taq DNA polymerase (639208, Clontech-Takara). The first PCRs contained 10âµg of genomic DNA per PCR, and the total reactions resulted in targeted amplification from one-third of the total genomic DNA. The first 16 cycles targeted PCR amplification and utilized the following primer set: mTKOv3-PCR1-F: ATTAGTACAAAATACGTGACGTAGAA (forward) and mTKOv3-PCR1-R: ACCTTCTCTAGGCACCGGATCA (reverse). The second PCRs were performed for 14 cycles using the following primers with adapters optimized to introduce the specific adapters for Illumina next-generation sequencing technology specific for the Hiseq4000: mTKO-P2: AATGATACGGCGACCACCGAGATCTACACGAGATCGGACTATCATATGCTTACCGTAACTTGAA (forward) and mTKO-P7##-IND: CAAGCAGAAGACGGCATACGAGATGCACGACGAGACGCAGACGAAnnnnnAGAGCAACTTCTCGGGGACTGTGGGCGA. Amplified PCR products from two replicates of the second PCR were pooled together and extracted from the agarose gel with the QIAquick gel extraction kit (28704, Qiagen). Samples were quantified using Qubit 2.0 DNA HS Assay (Q32851, Thermo Fisher), QuantStudio 5 System (Applied Biosystems) and Tapestation High Sensitivity D1000 Assay (5067-5582, Agilent Technologies). Six samples were pooled equilmolar to be run on a Nextseq 500 high-output 75-bp SR with 10% PhiX. Custom primers were required for read 1 (20ânt): mTKO-Seq-26bp TCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCG; and to obtain the sample index, read 2 (6ânt): mTKO-Seq-Index-7 AGATGCACGACGAGACGCAGACGAA.
CRISPR screen analysis
Bowtie63 was used to obtain raw read-counts with 1 mismatch allowance, thus taking the best-matching single guide RNA per read. Bayesian Analysis of Gene EssentiaLity 2 (BAGEL2) software64 was then used to calculate normalized read counts, and log2 fold change was obtained by comparing the reference timepoint of the corresponding cell line. Next, genes were determined as vulnerabilities by applying the standard BAGEL2 pipeline and excluding core-essential genes. The top 500 gene vulnerabilities ranked by BAGEL score were used as an input for enrichment pathway analysis using the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt)65,66. The following parameters were used for WebGestalt: minimum number of genes per categoryâ=â5; maximum number of genes per categoryâ=â2,000; and test performedâ=âBenjaminiâHochberg.
Statistical analysis and reproducibility
Samples were analysed as described in the figure legends. Data were considered significant if Pâ<â0.05. No statistical methods were used to predetermine sample size. For randomization and blinding details, please see the Reporting Summary. Most experiments were not randomized because they did not have an intervention that required randomization. Statistics were calculated using Graphpad Prism v9.0.1 software or RStudio 4.0.2 unless described otherwise in the Methods; statistical details can be found in the legends for each figure. The box for all box and whisker plots represent the 25th to 75th percentiles for minima and maxima, the line in the middle of the box represents the median, and the whiskers represent minimum and maximum values. Adobe Illustrator v26.3.1 was used to create the schematics and figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.