Mice
SPF C57BL/6J (B6) mice (Jax, catalogue no. 000664, both sexes) were obtained from The Jackson Laboratories. CD45.1 congenic mice (B6.SJL-Ptprca Pepcb/BoyJ, JAX, catalogue no. 002014), Rosa-CAG-LSL-tdTomato reporter mice (B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, JAX, catalogue no. 007908), Il17a-Cre mice (STOCK Il17atm1.1(icre)Stck/J, JAX, catalogue no. 016879) and ROSA26-eGFP-DTA (B6.129S6(Cg)-Gt(ROSA)26Sortm1(DTA)Jpmb/J, JAX, catalogue no. 032078) were purchased from The Jackson Laboratory. TCR7B8 (C57BL/6-Tg(Tcra,Tcrb)2Litt/J) and TCRHh7-2 (C57BL/6-Tg(Tcra,Tcrb)5Litt/J) mice were generated in house as described previously. All transgenic lines were bred and maintained under SPF conditions at the Alexandria Center for Life Sciences animal facility, New York University School of Medicine.
For IL-17A fate mapping experiments, sex-matched littermates (both male and female) were used. Experimental cohorts were 6–8 weeks old at treatment onset. Sample sizes were determined by power analysis (power = 0.9, α = 0.05) using mean and s.d. estimates from previous and pilot studies (four to five animals per group). All animal procedures were conducted in compliance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of New York University School of Medicine.
Antibodies, intracellular staining and flow cytometry
Monoclonal antibodies were obtained from eBioscience, BD Pharmingen, BioLegend, Thermo Fisher, Tonbo Bioscience and Invitrogen. The following fluorochrome-conjugated antibodies were used: CD4 BUV395 (GK1.5, BD, catalogue no. 563790, 1:400), CD25 APC (PC61, Thermo, catalogue no. 17-0251-82, 1:400), CD69 PE-Cy7 (H1.2F3, BioLegend, catalogue no. 104512, 1:200), CD44 AF700 (IM7, BD, catalogue no. 560567, 1:200) or BV510 (IM7, BD, catalogue no. 563114, 1:200), CD45.1 BV650 (A20, BD, catalogue no. 563754, 1:400), CD45.2 FITC (104, eBioscience, catalogue no. 11-0454-85, 1:400), CD19 PerCP-Cy5.5 (1D3, Tonbo, catalogue no. 65-0193-U100, 1:400), B220 PerCP-Cy5.5 (RA3-6B2, Invitrogen, catalogue no. 45-0452-82, 1:400), CD11c PerCP-Cy5.5 (N418, Invitrogen, catalogue no. 45-0114-82, 1:400) or PE-Cy7 (N418, BioLegend, catalogue no. 117318, 1:400), CD11b PerCP-Cy5.5 (M1/70, Invitrogen, catalogue no. 45-0112-82, 1:400) or BUV395 (BD, catalogue no. 563553, 1:400), MHCII I-A/I-E PerCP-Cy5.5 (M5/114.15.2, BioLegend, catalogue no. 107626, 1:400), NK1.1 PerCP-Cy5.5 (PK136, Invitrogen, catalogue no. 45-5941-82, 1:200), TCRβ BV711 (H57-597, BD, catalogue no. 563135, 1:200), TCRγδ PerCP-Cy5.5 (GL3, BioLegend, catalogue no. 118117, 1:400), FOXP3 FITC (FJK-16s, eBioscience, catalogue no. 11-5773-82, 1:200), RORγt BV421 (Q31-378, BD, catalogue no. 562894, 1:200), T-bet PE-CF594 (O4–46, BD, catalogue no. 562467, 1:70), IL-17A AF700 (TC11-18H10.1, BioLegend, catalogue no. 506914, 1:200), IFN-γ PE-Cy7 (XMG1.2, BioLegend, catalogue no. 505826, 1:200), Granzyme B AF700 (QA16A02, BioLegend, catalogue no. 372222, 1:200), TNF BV650 (MP6-XT22, BioLegend, catalogue no. 506333, 1:200), CXCR6 PE/Dazzle594 (SA051D1, BioLegend, catalogue no. 151117, 1:200), CD62L PE (MEL-14, BD Pharmingen, catalogue no. 553151, 1:400), and TCR Vβ14 FITC (14-2, BD Pharmingen, catalogue no. 553258, 1:400). Dead cells were excluded using 4′,6-diamidino-2-phenylindole (Sigma) or LIVE/DEAD Fixable Blue dye (Thermo Fisher).
For scTCR-seq coupled with scRNA-seq, cells were labelled with TotalSeq-C hashtag antibodies (BioLegend): Hashtag 1 (M1/42; 30-F11, catalogue no. 155861, 1:100), Hashtag 2 (catalogue no. 155863, 1:100), Hashtag 3 (catalogue no. 155865, 1:100), and Hashtag 4 (catalogue no. 155867, 1:100).
For transcription factor staining, cells were first stained for surface markers, then fixed and permeabilized using the FOXP3 staining buffer set (eBioscience), followed by nuclear staining. For intracellular cytokine analysis, cells were stimulated for 3 h in RPMI-1640 culture medium supplemented with 10% fetal bovine serum (FBS), plus phorbol 12-myristate 13-acetate (50 ng ml−1, Sigma), ionomycin (500 ng ml−1, Sigma) and GolgiStop (BD Biosciences), then stained for surface markers, fixed, permeabilized and subjected to intracellular/nuclear staining with eBioscience buffers.
Flow cytometry was performed using BD LSR II or Aria II instruments (BD Biosciences), data acquisition was carried out using BD FACSDiva software (v.8.0.1; BD Biosciences) and data were analysed with FlowJo software (v.10.10.0) (Tree Star).
Design of SFB-3340 antigen construct and generation of cancer cell lines expressing SFB-3340
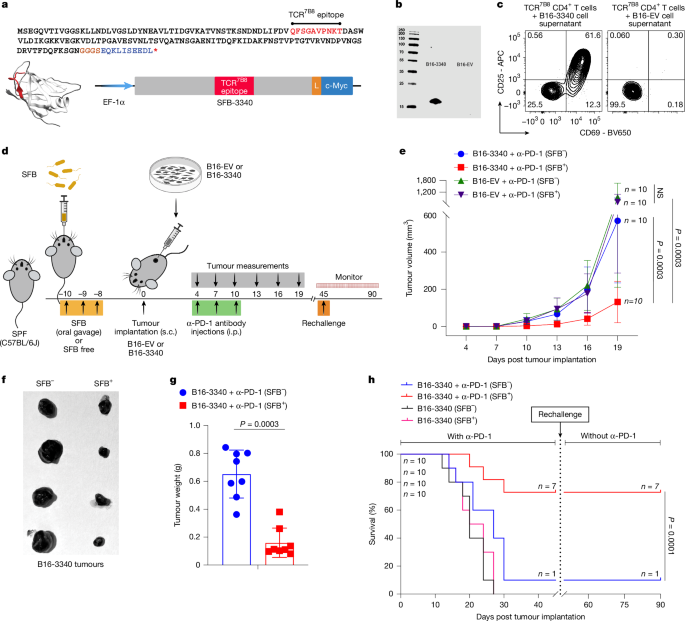
To establish a synthetic neoantigen mimicry model, we designed a construct encoding a small, independently folded domain containing a well-characterized immunogenic CD4+ T cell epitope (hereafter referred to as SFB-3340) derived from a large membrane protein of SFB (SFBNYU_003340, GenBank: EGX28318.1)36. A mammalian codon-optimized gene fragment encoding SFB-3340 antigen, fused via a flexible linker to a c-Myc tag, was synthesized chemically (GenScript) and cloned into the pEF1α-IRES-Neo vector (Addgene, catalogue no. 28019) between NheI and SalI restriction sites. Expression was driven by the constitutive EF1α promoter.
To establish stable B16-F10 and LLC1 cell lines expressing the neoantigen construct SFB-3340 (referred to as B16-3340 and LLC1-3340, respectively), B16-F10 cells (ATCC, catalogue no. CRL-6475) and LLC1 cells (ATCC, catalogue no. CRL-1642) were transfected with the expression plasmid complexed with TransIT-293 transfection reagent (Mirus Bio). The following day (approximately 18–22 h after transfection), the culture medium (DMEM supplemented with heat-inactivated 10% (v/v) FBS, 100 U ml−1 penicillin and 0.1 mg ml−1 streptomycin) was replaced with selection medium, which is same culture medium containing 1 mg ml−1 Neomycin (G-418) for selection. The cell cultures were then incubated for an additional 4–5 days, with the selection medium changed every 2 days to select for stably transfected clones. Single clones were isolated and further expanded in selection medium. The cells were passaged several times before assessing antigen expression by western blot and ex vivo activation assays. As a control cell lines, B16-F10 and LLC1 cells were transfected with the empty vector (pEF1α-IRES-Neo without the SFB-3340 gene fragment) (referred to as B16-EV and LLC1-EV respectively), and also selected in 1 mg ml−1 G-418-containing medium following the same protocol as for the B16-3340 and LLC1-3340 cell lines.
For generation of MC-38 cells expressing SFB-3340 antigen, the coding sequence of the designed SFB-3340 construct was cloned into an MSCV-IRES-Thy1.1 retroviral vector. Retrovirus was produced by transient transfection of Plat-E packaging cells; viral supernatants were collected, filtered and used to transduce MC-38 cells in the presence of 8 µg ml−¹ polybrene with spinoculation. At 72 h after transduction, Thy1.1+ cells were purified by FACS, expanded and maintained at >95% Thy1.1+.
Immunoblotting and ex vivo activation assay
To confirm stable expression of SFB-3340 in B16-3340, LLC1-3340 and MC-3340 cells, and to test whether the expressed antigen could stimulate SFB-3340 antigen-specific TCR7B8 CD4+ T cells ex vivo36, antigen-expressing and empty vector control cell lines were lysed in M-PER reagent (Thermo Fisher Scientific) supplemented with a protease inhibitor cocktail (Complete Mini EDTA-free; Roche). Lysates were centrifuged at 17,000g to pellet cellular debris, and supernatants were stored at −20 °C.
For western blotting, normalized amounts of protein from the resulting cell lysates were resolved by SDS–PAGE and transferred to nitrocellulose membrane using the iBlot 2 Dry Blotting System (Invitrogen). Membranes were blocked in PBS blocking buffer (LI-COR) and incubated overnight with anti-c-myc antibody (1:2,000, dilution, Cell Signalling) at 4 °C. Next day, after washing in PBST (PBS and 0.1% Tween-20), membranes were incubated with fluorescently conjugated secondary antibodies (LI-COR) at 1:10,000 dilution in PBS blocking buffer for 1 h at room temperature and imaged using the LI-COR Odyssey CLx Imaging system in the 800-nm channel (LI-COR).
For the ex vivo activation assay, spleens from female SPF (SFB-free) CD45.2 mice (6–7 weeks, Jackson Laboratories) were dissociated into single-cell suspension using the GentleMACS Spleen Dissociation Kit (Miltenyi Biotec) and DCs were isolated with CD11c microbeads (Miltenyi). Naive antigen-specific CD4+ T cells were purified from spleens and lymph nodes of CD45.1 TCR7B8 transgenic mice by mechanical dissociation. Red blood cells were lysed using ACK lysis buffer (Lonza). Naive TCR7B8 CD4+ T cells were sorted as CD4+TCRβ+CD44loCD62LhiCD25−Vβ14+ using a FACSAria II (BD Biosciences).
In 96-well round-bottom plates (CELLTREAT), 2 × 104 DCs were incubated in RPMI + 10% FBS for 2 h at 37 °C and 5% CO2 with one of the following: 10 µl PBS, 10 µl of cell lysates either from 1 × 106 B16-3340, LLC1-3340 or MC-3340 cells, 10 µl of cell lysates from corresponding empty vector control cells or 500 nM of chemically synthesized SFB-3340 peptide (GenScript). After the 2-h incubation (antigen loading), 1 × 105 naive TCR7B8 CD4+ T cells were added to each well and co-cultured for an additional 20–24 h. T cell activation was assessed by flow cytometry based on CD69 and CD25 expression.
Colonization of mice with SFB by oral gavage
SFB colonization was achieved through three consecutive oral gavages using faecal pellets from SFB mono-associated mice, following previously described methods36,53. Briefly, fresh faecal pellets were homogenized through a 100-μm filter, pelleted at 3,400 rpm for 10 min, and re-suspended in PBS. Each animal was administered one-quarter pellet by oral gavage. Colonization was confirmed by quantitative PCR (qPCR) with SFB-specific primers using universal 16S primers as control. Primers used were: 16S F, CGGTGAATACGTYCGG; 16S R, GGWTACCTTGTTACGACTT54; SFB F, GACGCTGAGGCATGAGAGCAT; SFB R, GACGGCACGGATTGTTATTCA.
Hh culture and oral infection
Hh was provided by J. Fox (MIT) and cultured as described previously45. Frozen Hh stocks were maintained in Brucella broth with 20% glycerol at −80 °C. For culture, bacteria were streaked onto blood agar plates (Thermo Scientific Blood Agar with 5% sheep blood; Thermo Fisher) and incubated at 37 °C in a hypoxia chamber (Billups–Rothenberg) under a micro-aerobic atmosphere (80% N2, 10% H2, 10% CO2; Airgas) adjusted to 3–5% O2. After 4 days, bacteria were harvested with a pre-moistened sterile cotton swab, re-suspended in Brucella broth and administered to mice at a density of 1 × 108 colony-forming units of Hh (equivalent to around 1 optical density unit) by oral gavage. A second inoculation was performed 3 days later. Colonization was confirmed by qPCR with Hh-specific primers using universal 16S primers as control. Primers used were: 16S F, CGGTGAATACGTYCGG; 16S R, GGWTACCTTGTTACGACTT54; Hh F, CAACTAAGGACGAGGGTTG; Hh R, TTCGGGGAGCTTGAAAAC.
In vivo tumour models and antibody treatments
SPF female C57BL/6J mice (6–7 weeks; Jackson Laboratories) were either maintained SFB-free (SPF, SFB−) or colonized with SFB by oral gavage as described above36,53. For subcutaneous tumour studies, B16-F10, LLC1 and MC-38 cell lines expressing the SFB-3340 protein fragment (B16-3340, LLC1-3340, MC-3340) or matched empty vector control cells were cultured in complete growth medium and 200 μg ml−1 of G-418 was added during maintenance of the B16-F10 and LLC1 transductants. Tumour cells, harvested freshly at 50–60% confluence after three to four passages, were washed and re-suspended in sterile PBS. Mice were inoculated subcutaneously in the right flank with 2.5 × 105 cells in 100 μl PBS or with 5.0 × 105 cells in 100 μl PBS for LLC1 and MC-38 (day 0).
For checkpoint blockade (in vivo anti-PD-1) experiments, mice received 100 μl intraperitoneal (i.p.) injections of 250 μg of anti-PD-1 antibody (clone RMP1-14, BioXCell, catalogue no. BP0146) diluted in 1× PBS when tumours were palpable (for B16-F10 on days 4, 7 and 10; for LLC1 days 7, 10 and 13; and for MC-38 on days 5, 8 and 11) post tumour implantation. Tumour growth was monitored by caliper measurements, and tumour volume was calculated using the ellipsoid volume formula (0.5 × D × d2, where D represents the longer diameter and d is the shorter diameter). Sample sizes were not predetermined but were based on standards commonly practiced in the field. Allocation to experimental groups was random. To minimize microbiota-related variability, control mice were from the same litter, of the same sex, and housed in the same room. Experiments were conducted blinded where feasible. For some tumour studies, investigators responsible for SFB colonization and tumour measurement remained blinded until study completion. Blinding was not possible in some experiments owing to the risk of SFB cross-contamination. Mice were humanely euthanized if tumours reached a volume of 2,000 mm³ or if any signs of discomfort were observed by investigators or identified by the animal care staff, in accordance with institutional IACUC guidelines and daily monitoring.
For in vivo depletion of CD4+ and CD8+ T cells, mice received i.p. injections of 200 μg of either anti-CD4 (clone GK1.5, BioXCell, catalogue no. BE0003-1) or anti-CD8a (clone 2.43, BioXCell, catalogue no. BE0061) antibody per mouse. Injections were initiated 2 days before tumour implantation and continued twice weekly thereafter until experimental end-points. Control mice were injected with PBS. In parallel, mice received three i.p. injections of anti-PD-1 antibody (250 μg per mouse) on days 4, 7, and 10 post tumour implantation as described above. The depletion efficiency was >95% in all the mice as monitored by flow cytometry of peripheral blood/spleen.
Isolation of lymphocytes from tumour, intestinal tissues and lymphoid organs
For tumour-infiltrating lymphocyte isolation, tumours were harvested 17–20 days post-implantation, minced and digested in RPMI containing collagenase type 1 (250 U ml−1; STEMCELL Technologies), DNase I (100 μg ml−1; Sigma), dispase (0.1 U ml−1; Worthington) and 10% FBS with constant stirring at 37 °C for 30 min. The resulting cell suspension was filtered, and lymphocytes were isolated using a 40%/80% Percoll density gradient (GE Healthcare) and centrifuged at 800g for 20 min without brake. Cells at the interface were collected for downstream analysis.
For isolation of lymphocytes from the SILP and LILP, the entire small intestine or colon were dissected from mice. Mesenteric fat and Peyer’s patches were removed carefully from these tissues. Intestinal tissues were opened longitudinally, washed thoroughly to remove faecal matter and treated sequentially with 1× Hank’s Balanced Salt Solution containing 1 mM dithiothreitol at 37 °C for 10 min with gentle shaking (200 rpm), followed by two incubations in 5 mM EDTA at 37 °C for 10 min each to remove epithelial cells. The remaining tissues were then minced with scissors and digested in RPMI containing 10% FBS, dispase (0.05 U ml−1; Worthington), collagenase II (1 mg ml−1; Roche) and DNase I (100 μg ml−1; Sigma) at 37 °C for 45 min with constant shaking (175 rpm). The digested tissues were then filtered through a 70-μm strainer to remove large debris. Viable lamina propria lymphocytes were collected at the interface of a 40%/80% Percoll/RPMI gradient (GE Healthcare). For isolation of cells from lymph nodes and spleens, tissues were dissociated mechanically with the plunger of a 1 ml syringe and filtered through 70-μm cell strainers. Red blood cells were lysed with ACK buffer (Thermo Fisher) before downstream applications45.
MHCII tetramer production and staining
Fluorophore phycoerythrin (PE) and allophycocyanin (APC) conjugated, I-Ab/3340-A6 (SFB-peptide-specific) and I-Ab/HH-E2 (Hh_1713-E2 peptide-specific) MHCII tetramers were synthesized at the NIH tetramer core facility55. In brief, immunodominant epitopes QFSGAVPNKT (3340-A6) and QESPRIAAAYTIKGA (HH_1713-E2), validated with the corresponding hybridoma (TCR7B8 and TCRHh7-2 respectively) stimulation assay, were covalently linked to I-Ab via a flexible linker to produce pMHCII monomers. Soluble monomers were purified, biotinylated and tetramerized with PE- or APC-labelled streptavidin36. Analysis of tetramer+ cells was performed as previously described with minor modifications56. Briefly, cells were first re-suspended in FACS buffer with FcR block (anti-mouse CD16/32), 2% mouse serum and 2% rat serum. Cells were then stained with PE- and APC-conjugated tetramers (10 nM) at room temperature for 1 h in the dark. Subsequently, the cells were washed and subjected to antibody staining against surface molecules at 4 °C.
IFNγ ELISPOT assay
IFNγ ELISPOT assay was performed using a mouse IFNγ ELISPOT kit (R&D systems) according to the manufacturer’s instructions. Briefly, 5 × 104 CD4+ T cells, extracted and sorted from either B16-3340 tumour tissue or SILP of SFB+ and SFB− mice as described above, were stimulated with either SFB-3340 peptide (specific peptide) or Hh7-2 peptide (non-specific peptide). CD11c+ APCs (2 × 104), purified from the spleen of SPF (SFB-free) mice as described above, were used for antigen presentation in this assay. Dots (IFNγ producing cells) were enumerated automatically using ImmunoSpot software (v.5.0).
Adoptive transfer of naive TCR7B8 and TCRHh7-2 CD4+ T cells
Spleens were harvested from donor TCR7B8 tdTomato-ONΔIL-17a or TCRHh7-2 tdTomato-ONΔFoxp3 mice, disassociated mechanically, and treated with ACK lysis buffer (Lonza) to remove red blood cells. Naive CD4+ T cells (TCR7B8 or TCRHh7-2) were sorted by flow cytometry (FACS Aria II, BD Biosciences) based on the following surface markers: CD4+CD3+CD44loCD62LhiCD25−TCRVβ14+ (7B8) or TCRVβ6+ (Hh7-2). Sorted cells were then re-suspended in PBS on ice and injected intravenously (i.v.) into the tail vein in congenic isotype-labelled recipient mice colonized with SFB or Hh. Cells from indicated tissues were analysed 5 weeks post-transfer.
scRNA-seq and scTCR-seq experiment
scRNA-seq and scTCR-seq were performed using the Chromium Single Cell 5′ v.2 reagent kit and Chromium Single Cell Mouse TCR Amplification Kit (10x Genomics). Tumour-infiltrating lymphocytes from B16-3340 tumours and lymphocytes from the SILP of SFB− and SFB+ mice (n = 5 in each group) were isolated as described above. CD4+ T cells were then sorted from pooled cells of either SFB− or SFB+ tumour tissues or SILP of individual mice using FACS Aria II (BD Biosciences). Sorted CD4+ T cells from each group (Tumour SFB−, Tumour SFB+, SILP SFB− and SILP SFB+) were resuspended in PBS containing 0.05% BSA and stained with cell hashing antibodies, TotalSeq-C0301 to C0304 (BioLegend, catalogue nos. 155861, 155863, 155865 and 155867)57 for 20 min on ice. Cells were then washed three times with MACS buffer. CD4+ T cells from both SFB− and SFB+ tumours were combined at a 1:1 ratio. Similarly, CD4+ T cells from the SILP of SFB− and SFB+ mice were combined at a 1:1 ratio. Approximately 1.5 × 104 cells per sample were loaded onto the Chromium Controller (10x Genomics) and libraries were prepared with the Chromium Single Cell 5′ kit following the manufacturer’s instructions. Libraries were sequenced using the NovaSeq 6000 system with a sequencing depth of more than 20,000 paired-end reads per cell. Sequencing reads were aligned to the mouse reference genome (mm10-2020-A, 10x Genomics) using Cell Ranger (v.7.1.0; 10x Genomics). Downstream data were processed and analysis were performed with the R packages Seurat v.5.1.0 (ref. 58).
Data processing of scRNA-seq
To preprocess single-cell data, raw scRNA-seq data were processed with Cell Ranger ‘multi’ software (v.7.1.0, 10x Genomics) using the mouse reference genome (mm10 2020-A, 10x Genomics). For scTCR-seq, data were aligned and quantified with CellRanger ‘multi’ software (v.6.6.1, 10x Genomics) against the reference vdj_GRCm38_alts_ensembl-5.0.0, using default parameters.
For scRNA-seq analysis, cells with fewer than 200 detected genes or more than 5% mitochondrial gene content were excluded. Hashtag oligonucleotides (HTO) counts were normalized using centred log ratio transformation and demultiplexing with Seurat::HTODemux function (positive quantile set to 0.99). Doublets mapped to several HTO tags were removed. RNA counts were normalized with Seurat::SCTransform function, regressing out cell cycle, ribosomal and mitochondrial scores59. Paired SFB− and SFB+ samples from the same tissue were integrated using the Seurat standard scRNA-seq integration workflow with 3,000 anchor genes. A shared nearest neighbour graph was constructed using the first 40 principal components and Leiden clustering (Seurat::FindClusters function) was applied at several resolutions to identify potential rare subsets60. Clusters were annotated based on canonical markers and differentially expressed genes identified with Seurat::FindAllMarkers (logistic regression model). Cells were then projected onto a UMAP for visualization61.
TCR sequence data were processed using Cell Ranger vdj pipeline to identify TCR genes and CDR3 sequences. For each sample, full length, productive TRB and TRA chains were retained for downstream analysis. Clonal expansions were defined as clonotypes with identical CDR3 nucleotide sequences of both chains present in at least three cells across all samples. TCR metadata were merged with the scRNA-seq Seurat object by cell barcodes and sample ID. Phenotypic characterization of TCR clonotypes was performed by exporting metadata from the Seurat object and analysed and quantified in Microsoft Excel (v.16.73).
Differential gene expression between groups was tested with the MAST package (MAST_1.28.0) as implemented in Seurat v.5.1.0 (ref. 62), which applies to hurdle model adapted to scRNA-seq data. Genes with Bonferroni-adjusted P value < 0.05 were considered as statistically significant.
Statistical analysis
Statistical tests including unpaired two-sided t-test, paired two-sided t-test, one-way ANOVA with Bonferroni correction, two-way ANOVA with Sidak’s multiple comparisons, Mann–Whitney test and the Mantel-Cox test for survival curves were all performed to compare the results using GraphPad Prism v.9 (GraphPad). No samples were excluded from analysis. Exact P values are reported where possible, and P < 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.