Mouse models
Housing conditions
All animal experiments in this study were performed in accordance with protocols approved by the Memorial Sloan Kettering Institutional Animal Care and Use Committee (approval number: 11-06-012). The mice were housed with a 12-h light–dark cycle between 8:00 and 20:00 in a temperature-controlled room (22 ± 1 °C) with free access to water and food. Both male and female mice were used in equal proportions for all experiments. No sex-based differences were observed. Experiments were performed using mice aged 10–14 weeks. Sample sizes were determined on the basis of previous experiments and published studies to ensure adequate power to detect biologically relevant differences. Mice were randomly assigned to experimental groups. Investigators were blinded to group allocation during data collection and analysis whenever possible.
Generation of an inducible Ogdh-knockdown mouse
Considering that the Ogdh knockout is embryonic lethal51, we developed an inducible model using doxycycline-inducible shRNAs linked to a GFP reporter. This system enables temporal and reversible suppression of Ogdh expression and facilitates the tracing and analysis of cells with Ogdh knockdown using the GFP reporter. To account for potential off-target effects of RNA interference (RNAi), we used two validated shRNAs (shOgdh_2081 and shOgdh_346)1. As a control for non-sequence-based effects of perturbing the RNAi machinery, we used a similar construct containing a Renilla-luciferase-targeting shRNA (shRen_731), which does not target any gene expressed in mouse cells. The guide strands for shRNAs are: Renilla, TAGATAAGCATTATAATTCCT; Ogdh_2081, TAAATGAAACATTTTGTCCTG; Ogdh_346, TAGCAATTCTGCATACTTCTG. Doxycycline-inducible GFP-coupled shRNA constructs were electroporated and integrated into a ‘homing cassette’ at the Col1a1 locus in 4482 ES cells30,33; this cassette contains a doxycycline-inducible reverse Tet transactivator (rtTA-M2) expressed from the Cag-rtTA promoter. Validated clones were used to generate chimeric mice using eight-cell aggregation, allowing the assessment of functionality in F0. The resulting founders were backcrossed to establish germline transmission, generating Tg. TRE–shOgdh_2081, Tg. TRE–shOgdh_346 and Tg. TRE–shRenilla_731 mice. To further amplify Ogdh knockdown and enable widespread expression of the TRE-GFP-shRNA cassette, including in the intestine33, we crossed TRE–shRenilla and TRE–shOgdh mice with CAGs-rtTA3 transgenic mice, resulting in TRE–shRenillaCag-rtTA3 and TRE–shOgdhCag-rtTA3 mice.
ISC analysis
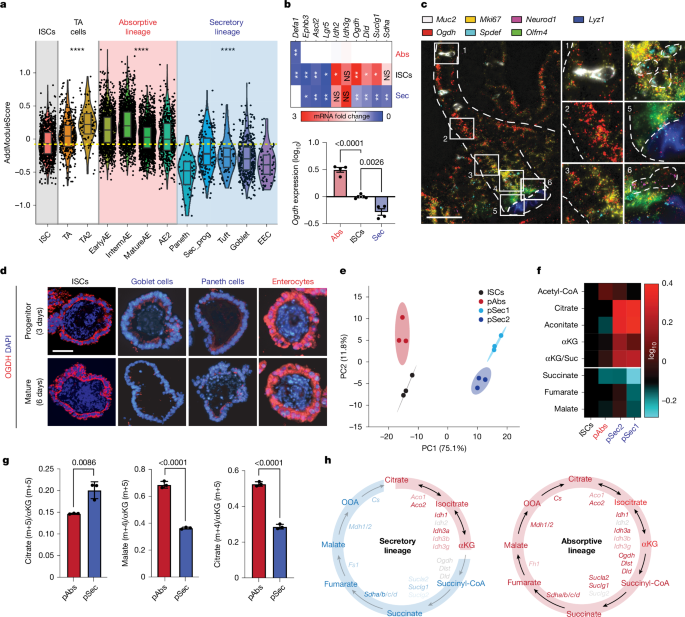
To enable isolation of LGR5+ cells for assessing the expression of TCA-cycle enzymes, we used Lgr5-EGFP-IRES-creERT2 mice52.
Generation of a reporter mouse for secretory progenitors
To address whether the secretory pool has plasticity and could undergo dedifferentiation into ISCs, a reporter mouse was generated by crossing the Atoh1-creERT2 mouse model47 with Rosa26-CAGs-LSL-RIK53, which contains a loxP-flanked stop cassette upstream of the RtTA3, an IRES sequence and the monomeric far-red fluorescent protein mKate2, all inserted into the Gt(ROSA)26 locus. Administration of 4-hydroxytamoxifen leads to the excision of the stop cassette in the RIK allele. Consequently, ATOH1+ cells and their progeny will permanently express mKate2, enabling their identification and the dynamic tracking of the fate of ATOH1-expressing cells.
Generation of an immune-mediated colitis model (Rag2
−/− mice)
To generate a rodent model of human inflammatory bowel disease, we used the CD4+CD45RBhigh-induced colitis model in Rag2−/− mice45. In brief, spleens from ten C57Bl/6 male mice were collected, smashed, filtered through a 40-μm filter and washed with isolation buffer (phosphate-buffered saline (PBS), 0.5% bovine serum albumin (BSA) and 2 mM EDTA, pH 7.2). The cells were then centrifuged (288g, 5 min), and the resulting splenocyte pellets were resuspended in ACK buffer (Quality Biologicals, 118-156-101CS) to lyse red blood cells. After cell counting, CD4+ cells were isolated using the CD4+ isolation kit (Miltenyi, 130-104-454) following the manufacturer’s instructions. The splenocytes were transferred to fluorescence-activated cell sorting (FACS) buffer (0.5% BSA and 2 mM EDTA in Ca2+/Mg2+-free PBS) and incubated on ice for 30 min with anti-CD4–APC (BioLegend, 116014, clone RM4-4, 1:200) and anti-CD45Rb–FITC (BioLegend, 103306, clone C363-16A, 1:200) antibodies. CD4+CD45Rbhigh and CD4+CD45Rblow cells were then sorted using a Sony MA900 cell sorter. Finally, 0.5 × 106 cells were intraperitoneally injected into Rag2−/− male mice, and colitis developed within the following three months.
Mouse diets and treatments
Cag-rtTA3 mediating shRenilla and shOgdh expression was activated by feeding mice with a doxycycline hyclate diet (200 mg kg−1) at adult stage. The food was changed twice per week.
Acute DSS treatment was performed as previously described43. In brief, at ten weeks of age, mice were treated with 2% DSS (MP-Biomedicals, 021160110-CF) in drinking water for five days. Afterwards, the water was changed to regular drinking water and mice were euthanized at the indicated time points. In all experiments with DSS models, mice were weighed at the beginning of DSS treatment and every other day thereafter. In addition, they were evaluated daily for signs of distress or end-point criteria. Specifically, mice were immediately euthanized if they lost more than 20% of their initial body weight or showed breathing difficulties.
For the bromodeoxyuridine (BrdU) pulse experiment, mice were injected with 1 mg of BrdU (Sigma-Aldrich) in PBS and euthanized two hours later.
For DM-αKG supplementation, mice were injected (intraperitoneally) once daily with 600 mg kg−1 or 300 mg kg−1 of DM-αKG (349631-5G, Sigma-Aldrich) dissolved in PBS. For the vehicle control, mice were injected with PBS.
For pulsatile inhibition of Ogdh, mice were injected intraperitoneally with doxycycline (2.5 mg kg−1, Sigma-Aldrich, D9891) once daily in cycles of three consecutive days on followed by four days off.
DM-succinate treatment was performed as described54. In summary, mice were provided with 100 mM DM-succinate (W239607-1KG-K, Sigma-Aldrich) in their drinking water and received intraperitoneal injections of 100 mM DM-succinate every other day throughout the experiment. The pH was adjusted to 6.5 for both the drinking water and the injections.
For pulse-chase labelling experiments in Atoh1-creERT2;RIK mice, 5 mg of tamoxifen (T5648-1, Sigma-Aldrich) per mouse was administered by oral gavage every other day for a total of five days.
For glycolysis inhibition, mice were treated with intraperitoneally injected with 500 mg kg−1 of 2-deoxyglucose (Sigma, 111980050) five times per week for one month.
Genotyping PCR
Genomic DNA was extracted from mouse tails and ear punches. Biopsies were digested in MGB buffer supplemented with 10% Triton X-100, 1% 2-mercaptoethanol and 0.4 mg ml−1 proteinase K (Qiagen, 19133), and incubated overnight at 55 °C. PCR conditions were 95 °C for 6 min, then 35 cycles (95 °C for 40 s, 62 °C for 45 s, 72 °C for 1 min), then 72 °C for 10 min. PCR amplification yielded a 300-bp band for the mutant allele (indicating successful shRNA integration) and a 218-bp band for the wild-type allele.
RNA-seq analysis and qPCR
RNA extraction and RNA-seq library preparation and sequencing
Total RNA was isolated from crypts isolated from TRE-shRenillaCag-rtTA3, TRE-shOgdhCag-rtTA3, vehicle-treated or DM-αKG-treated mice using RNeasy kits (QIAGEN, 74004). RNA concentration and quality was assessed using an Agilent 2100 Bioanalyzer. Sequencing and library preparation was performed at the Integrated Genomics Operation at the Memorial Sloan Kettering Cancer Center (MSKCC). RNA-seq libraries were prepared from total RNA. After RiboGreen quantification and quality control by Agilent Bioanalyzer, 100–500 ng of total RNA underwent polyA selection and TruSeq library preparation according to the instructions provided by Illumina (TruSeq Stranded mRNA LT Kit, RS-122–2102), with eight cycles of PCR. Samples were barcoded and run on a HiSeq 4000 or HiSeq 2500 in a 50-bp/50-bp paired-end run, using the HiSeq 3000/4000 SBS Kit or TruSeq SBS Kit v4 (Illumina) at MSKCC’s Integrated Genomics Operation. An average of 41 million paired-end reads was generated per sample. Ribosomal reads represented at most 0.01% of the total reads generated, and the fraction of mRNA averaged 53%.
RNA-seq read mapping, differential expression analysis and heat-map visualization
Adaptor sequences were removed from the RNA-seq data using Trimmomatic55. The trimmed reads were then aligned to the GRCm38.91 (mm10) reference genome using STAR56, and the transcript count was quantified using featureCounts57 to generate a raw count matrix. Differential gene expression analysis was performed using the DESeq2 package58 in R (http://cran.r-project.org/), comparing the experimental conditions. Each condition had three to five independent biological replicates (individual mice). PCA was performed using DESeq2 to visualize the variation in gene expression among the samples. Differentially expressed genes (DEGs) were identified on the basis of a greater than twofold change in gene expression with Padj < 0.05. To visualize the DEGs, the samples were z-score normalized and plotted as a heat map using the ‘pheatmap’ package in R. Functional annotations of the gene sets were done by pathway enrichment analysis using the Reactome, Azimut, CellType and KEGG databases. The analysis was performed with enrichR59, and the significance of the tests was assessed using a combined score, calculated as log(P) × z, where P represents the P value from the Fisher exact test, and z is the z-score indicating the deviation from the expected rank. Gene set enrichment analysis (GSEA)60 was done using the GSEA-Preranked tool (v.2.07). The analysis involved the enrichment of gene sets using RNA-seq data obtained from the experiment. The gene sets were derived from the MSigDB database (http://software.broadinstitute.org/gsea/msigdb), as well as previously published signatures from mouse intestine.
scRNA-seq data analysis
Data were obtained from a previously published study26,27,28. In brief, human intestines were collected from three male donors (aged 29, 45 and 53 years) through HonorBridge (formerly Carolina Donor Services). Donors met eligibility criteria, including the absence of infectious diseases, cancer or recent abdominal surgeries. Intestinal tissues were divided into six regions: duodenum, jejunum, ileum and the ascending, transverse and descending colon. Mucosectomies (3 × 3 cm) were taken from the centre of each region. Sample preparation and cell hashing were performed as described using the reported cell classification and anatomical location26,27,28. Raw and normalized data, along with cell annotations, were obtained from a published study and downloaded from GSE185224. Seurat60,61,62,63 was used to perform the scRNA-seq data analysis, and the.h5ad file was converted into a Seurat object using the R package zellkonverter. The GetAssayData function was used to extract Ogdh expression, and the AverageExpression function was used to calculate the average expression across different regions or lineages. The TCA-cycle gene set was generated using the genes from the TCA-cycle enzymes, and the gene signature score was computed using the AddModuleScore function from Seurat. Gene signatures used in the paper are shown in Supplementary Table 8.
qPCR with reverse transcription
For qPCR with reverse transcription (qRT–PCR) analysis, total RNA was extracted from mouse ES cells, isolated crypts or sorted cells from Lgr5-EGFP mice using the RNeasy Mini Kit (QIAGEN, 74004). Subsequently, cDNA synthesis was performed using TaqMan reverse transcription reagents (Applied Biosystems). qPCR was performed in triplicate using SYBR Green PCR Master Mix (Applied Biosystems) on the ViiA 7 Real-Time PCR System (Invitrogen). The expression levels of target genes were normalized to endogenous control genes, Rplp0 (also known as 36B4) and Actb. Gene-specific primer sets were designed using the qPrimerDepot tool provided by the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) or published elsewhere. For primer sequences and details, see Supplementary Table 2.
Bisulfite conversion, detection of 5mC and 5hmC loci, primer design and normalization
To quantify 5mC and 5hmC at SPDEF-specific loci, we used the BisulPlus Loci 5mC & 5hmC Detection PCR Kit (P-1067-48), following the manufacturer’s protocol. For DNA extraction and bisulfite conversion:, genomic DNA was extracted from intestinal crypts using the Zymo Research Quick-DNA Miniprep Plus Kit (D4069) and quantified with a NanoDrop spectrophotometer (Thermo Fisher Scientific). For each reaction, 500 ng genomic DNA was subjected to bisulfite conversion to deaminate cytosines while preserving 5mC and 5hmC. The conversion was performed according to the manufacturer’s guidelines. For primer design, primers for bisulfite-converted DNA were designed using MethPrimer (https://methprimer.com/cgi-bin/methprimer/methprimer.cgi/) (Supplementary Table 7). The primer design avoided CpG dinucleotides to minimize bias between methylated and unmethylated DNA. Primers were 26–35 bases long to ensure specificity, with melting temperatures (Tm) adjusted to higher than 60 °C through guanine-rich sequences. Each primer set amplified one strand of the bisulfite-converted DNA. For PCR amplification and detection of 5mC and 5hmC, after bisulfite treatment, the targeted loci were amplified in a 50-µl PCR reaction containing 25 µl of 2× Master Mix, 2 µl of bisulfite-converted DNA, 1 µl of each primer (10 µM) and nuclease-free water. The thermal cycling conditions were: initial denaturation: 95 °C for 5 min; 35 cycles of 95 °C for 30 s (annealing temperature specific to primers) and 72 °C for 1 min; final extension: 72 °C for 5 min. For normalization of 5hmC levels: to accurately quantify 5hmC, PCR signals from the 5hmC-specific reactions were normalized against the total methylation (5mC + 5hmC) at each locus. The percentage of 5hmC was calculated using the following formula: 5hmC (%) = 100 × (5hmC signal/(5mC signal + 5hmC signal)).
For qPCR, the Ct values of the 5mC- and 5hmC-specific reactions were compared, and the relative levels of 5hmC were obtained using the ΔCt method. The difference between Ct values (ΔCt) provided an estimate of relative hydroxymethylation, and the fold change was calculated using the formula 2−ΔCt2.
Genetic constructs and plasmids
For plasmid maps and shRNA sequences, see Supplementary Table 6.
The lentiviral vector used to silence Hnf4a and Hnf4g, pLV[2miR30]-Hygro-TRE3G>mCherry:{shHNF4a}:{shHNF4g}, and its control vector, pLV[2miR30]-Hygro-TRE3G>mCherry: Scramble [miR30-shRNA#1]:Scramble [miR30-shRNA#2], were constructed and packaged by VectorBuilder. These are tandem shRNA expression vectors, each carrying two shRNA sequences designed to be co-expressed under the same regulatory elements, enabling simultaneous knockdown of two target genes or, in the case of the control, two non-targeting scramble sequences. The vector IDs are VB231003-1530dwr (shControl vector) and VB231003-1530srj (Hnf4a and Hnf4g vector), respectively. These IDs can be used to retrieve specific information about the plasmids. shHnf4a: TACTATTTTACCTACCTATGGG; shHnf4g: TTAATATTTATGTCAGTGCTGG; shRenilla: ACCTAAGGTTAAGTCGCCCTCG (×2 tandem sequence).
The reporter vectors used to study the role of the HNF4 family in Ogdh expression, pRP[Pro]-hRluc/Puro-{Ogdh_Promoter_WT}>TurboGFP and pRP[Pro]-hRluc/Puro-{Ogdh_Promoter_Hnf4a_Mutant}>TurboGFP, were constructed by VectorBuilder. The vector IDs are VB231003-1479yab and VB231003-1483kbd, respectively. Wild-type Ogdh promoter: AACAAGTGTTCAAAATAGTCACTCATGTTATTCAAATTATTTTGTGCAGGGATACATTTTACCAACCCAACTATTATTTAGGGCAGCTTGTTTTGGATACAAAGCCCGTGGGCCTCAAAGTCGCAGCGCTCCTGCTTCGGCCCGCCAAACGCTTCAATTATCAGACGGCATCCCACGCCCTGAATGTACCAGGTTCTTAACAAGCTTCGGAAGCGTCTCCCGTGTAACTGCTAATGACAGCCGAAAGACAGTGAGCAACAGGCTGGCTTTGGCCAGATGCAAAGTTCTGCATTGGCGCGAAGCCCGAGCGAGCGACTGAAACCCAATTCTGTGACGTCACGTCACGCCCACAGCCTGTCTTGCAGGCCGCTCCTCTGGGGCCGGGCTACGCGTTGACGCC. Mutated Ogdh promoter: AACAAGTGTTCAAAATAGTCACTCATGTTATTCAAATTATTTTGTGCAGGGATACATTTTACCAACCCAACTATTATTTAGGGCAGCTTGTTTTGGATACAAAGCCCGTGGGCCTCAAAGTCGCAGCGCTCCTGCTTCGGCCCGCCAAACGCTTCAATTATCAGACGGCATCCCACGCCCTGAATGTACCAGGTTCTTAACAAGCTTCGGAAGCGTCTCCCGTGTAACTGCTAATGACAGCCGAAAGACAGTGAGCAACAGGCTGGGGGAGCTAGATGGGGCGATTTGCATTGGCGCGAAGCCCGAGCGAGCGACTGAAACCCAATTCTGTGACGTCACGTCACGCCCACAGCCTGTCTTGCAGGCCGCTCCTCTGGGGCCGGGCTACGCGTTGACGCC.
Intestine preparation
For immunofluorescence analysis
Small intestine and colon were removed from mice and flushed with PBS. Intestines were then opened longitudinally, ‘Swiss-rolled’, incubated overnight in 10% formalin at room temperature, changed to 70% ethanol and processed for paraffin embedding.
For isolation of crypts
From freshly dissected small intestines from C57Bl/6, TRE-shOgdhCag-rtTA3 and TRE-shRenCag-rtTA3 mice, the duodenum (first 10–12 cm from the stomach) was isolated, as it yields more crypts in a shorter time. The duodenum was longitudinally opened through the lumen, then washed in ice-cold Hanks’ balanced salt solution (HBSS) until the rest of the samples were collected. Next, the duodenum was transferred to a 15-ml Falcon tube, then chopped into pieces of 1–2 cm. For each tube, 6–7 ml of ice-cold 8 mM EDTA in HBSS was added, and the tube was incubated on ice for 15 min. After incubation, the EDTA solution was removed and replaced with HBSS. The tube was then vigorously shaken for approximately 20 s. A 20-μl aliquot was taken from the supernatant and examined under a microscope to confirm that it contained the villus fraction. The supernatant was discarded, and the intestine pieces were then transferred to a new 15-ml Falcon tube and rinsed in ice-cold HBSS to remove any residual villi. Another 6–7 ml of ice-cold 8 mM EDTA in HBSS was added per tube, followed by a 15-min incubation on ice. As in the previous step, the EDTA solution was removed and replaced with HBSS (optional step), and the tube was shaken vigorously for approximately 20 s. A 20-μl aliquot of the supernatant was taken and examined under the microscope to check whether it contained the desired crypt fraction. Depending on the yield, these steps were repeated until the crypts were enriched in the supernatant.
Culture of intestinal organoids
Regular conditions
Freshly isolated crypts were embedded in 50 μl of undiluted Matrigel (356231, Cultek) and cultured in DMEM/F-12 (D8437, Sigma-Aldrich) supplemented with penicillin–streptomycin, 1× (2 mM) Glutamax (35050038, Gibco or Life Technologies, Thermo Fisher Scientific), 10 mM HEPES (15630049, Gibco or Life Technologies, Thermo Fisher Scientific), 2 mM N-acetyl cysteine (A8199-10G, Sigma-Aldrich), 1× B27 supplement (17504-044, Life Technologies), 10 mM nicotinamide (N0636-100G, Sigma-Aldrich), 50 ng ml−1 recombinant mEGF (PMG8044, Gibco), 100 ng ml−1 recombinant Noggin (250-38, PeproTech), 1 μg ml−1 mouse R-spondin 1 (120-38, PeproTech) and 1% normocin (ant-nr-1, InvivoGen)64. During the initial 12-h culture establishment, 1.5 μM CHIR99021 (GSK3 inhibitor, 2520691; PeproTech) and 10 μM Y-27632 inhibitor (1293823, PeproTech) were included, and then the medium was replaced. The medium was changed every other day.
Organoid differentiation
Organoid differentiation assays were performed as previously described29. In brief, crypts or single cells were entrapped in Matrigel and plated at the centre of wells in a 24-well plate. After polymerization of Matrigel, 500 μl of complete Advanced DMEM/F-12 was added. The enrichment media were as follows. ISC enrichment medium (ENR-CV): EGF (50 ng ml−1, Life Technologies), Noggin (100 ng ml−1, PeproTech), R-spondin 1 (500 ng ml−1, R&D) and small molecules including CHIR99021 (3 μM, Stemgent) and valproic acid (1 mM, Sigma-Aldrich). Paneth cell enrichment medium (ENR-CD): EGF (50 ng ml−1, Life Technologies), Noggin (100 ng ml−1, PeproTech), R-spondin 1 (500 ng ml−1, R&D) and small molecules including CHIR99021 (3 μM, Stemgent) and DAPT (10 µM, Sigma-Aldrich). Goblet cell enrichment medium (ENR-VD): EGF (50 ng ml−1, Life Technologies), Noggin (100 ng ml−1, PeproTech), R-spondin 1 (500 ng ml−1, R&D) and small molecules including valproic acid (1 mM, Sigma-Aldrich) and DAPT (10 µM, Sigma-Aldrich). Enterocyte enrichment medium (ENR-IV): EGF (50 ng ml−1, Life Technologies), Noggin (100 ng ml−1, PeproTech), R-spondin 1 (500 ng ml−1, R&D) and small molecules including valproic acid (1 mM, Sigma-Aldrich) and IWP2 (2 μM, Sigma-Aldrich). To enrich in progenitor cells, organoids were kept in ENR-CV for six days and then transferred for three days to the corresponding differentiation medium. For fully mature lineages, ISC-enriched organoids were maintained in differentiation medium for at least six days.
Intestinal organoid engineering
Organoid nucleofection protocol
Freshly intestinal organoids were isolated as previously described. They were maintained in ENR medium with CHIR99021 (10 μM) for four days before electroporation. Five drops of Matrigel (30–40 μl) were plated per six-well plate, and the medium was refreshed the day before electroporation. On the day of electroporation, organoids were recovered from Matrigel using Cell Recovery Solution (Corning, 76332-050) and spun down at 135g for 5 min at 4 °C. The organoids were then resuspended in 200 μl TrypLE (Life Technologies, 12604021) per well and incubated for 3 min at 37 °C in a water bath. Subsequently, 5 ml PBS was added, and the organoids were further mechanically dissociated. For nucleofection, we used 2–3 μg of the desired plasmid; specifically, a reporter construct carrying the HNF4-binding site in the Ogdh promoter, either mutated or wild type. The P3 Primary Cell 4D-Nucleofector X Kit (Lonza, V4XP-3024) was used for nucleofection. The nucleofection buffer was prepared immediately before the procedure, following the manufacturer’s instructions. Organoids were resuspended in 20 μl nucleofection buffer and immediately subjected to electroporation using the ESC program in the 4D-Nucleofector (Lonza). After electroporation, 100 μl ENR medium with CHIR99021 and Y-27632 inhibitors was added to each well. The nucleofected organoids were transferred to an Eppendorf tube, spun down at 1,200 rpm for 5 min and resuspended in Matrigel. Organoids were allowed to recover overnight before experiments.
Organoid viral transduction protocol
Organoids were isolated as described above and maintained in ENR medium supplemented with CHIR99021 (10 μM) for four to six days. Next, double Hnf4a and Hnf4g shRNA along with their respective controls, were transduced by spinoculation, which was performed as previously described64. After spinoculation, organoids were spun down at 1,200 rpm for 5 min and resuspended in Matrigel. The organoids were then allowed to recover before selection or sorting.
Organoid treatments
For the experiments using organoids, the following treatments were applied: doxycycline at 2 μg ml−1 (Sigma-Aldrich, D9891-25G), DM-αKG at 3.35 mM (1:2,000 dilution from stock) (Sigma-Aldrich, 102418940), octyl-αKG at 10 mM final concentration (Sigma-Aldrich, 876150-14-0), octyl-L-2HG at 10 mM final concentration (Sigma-Aldrich, 1391194-64-1) and DM-succinate at 3 mM final concentration (Sigma-Aldrich, W239607).
Metabolomics
Intestinal organoid culture
Intestinal organoid culture was performed by isolating crypts through mechanical disruption with EDTA, followed by embedding them in Matrigel. To ensure consistent numbers of cells, crypts from five independent mice were pooled together, and the resulting pool was embedded in Matrigel. Five Matrigel drops (30–40 µl per drop) were plated in every six-well plate. Triplicate samples were plated from the pooled crypts to maintain consistency in the experiments, because the LC–MS is highly sensitive to variations in cell number, and it is challenging to plate the same number of organoids from different mice. Organoids were kept in the pertinent culture medium as described above. Also, as an internal control for the metabolites present in Matrigel, in LC–MS experiments, we isolated metabolites from Matrigel (same number of drops) without organoids but going through the same culture protocol. For isotopologue tracing, organoids were transferred to ENR-X medium containing either 13C6 glucose or 13C5 glutamine, without Glutamax, and incubated for 24 h before sample collection.
Sample preparation
Supernatant was aspirated from Matrigel cultures with or without organoids. The cells were lysed and metabolites were extracted by adding 1 ml ice-cold 80% methanol directly to the Matrigel drops, followed by incubation overnight at −80 °C to aid protein precipitation. The following day, the methanol extracts were centrifuged at 20,000g for 20 min at 4 °C, and 800 µl of the supernatant was transferred to a new tube and evaporated in a vacuum concentrator (GeneVac).
LC–MS/MS analysis
For metabolomic profiling using LC–MS, dried extracts were resuspended in 80 µl of 40% acetonitrile in water + 20 µl of 100% methanol for hydrophilic interaction liquid chromatography (HILIC) or in 100 µl of 50% methanol in water for ion-pair LC separations. Samples were vortexed and incubated on ice for 20 min, vortexing every 5 min to ensure adequate resuspension. All samples underwent one final centrifugation (20,000g for 20 min at 4 °C) to remove any residual particulate.
HILIC LC–MS analysis was performed on a 6545 Q-TOF mass spectrometer (Agilent Technologies) in positive ionization mode. LC separation was done on an ACQUITY UPLC BEH Amide column (150 mm × 2.1 mm, particle size 1.7 μm, Waters) using a gradient of solvent A (10 mM ammonium acetate in 10:90 acetonitrile: water with 0.2% acetic acid, pH 4) and solvent B (10 mM ammonium acetate in 90:10 acetonitrile: water with 0.2% acetic acid, pH 4). The gradient was 0 min, 95% B; 9 min, 70% B; 13 min, 30% B; 14 min, 30% B; 14.5 min, 95% B; 15 min, 95% B; and 20 min, 95% B. Other LC parameters were as follows: flow rate 400 μl min−1, column temperature 40 °C and injection volume 5 μl. Other MS parameters were as follows: gas temperature 300 °C, gas flow 10 l min−1, nebulizer pressure 35 psi, sheath gas temperature 350 °C, sheath gas flow 12 l min−1, Vcap 4,000 V and fragmentor 125 V.
Ion-pair LC–MS analysis was performed on a 6230 TOF mass spectrometer (Agilent Technologies) in negative ionization mode. LC separation was done on an XSelect HSS T3 column (150 mm × 2.1 mm, particle size 3.5 μm, Waters) using a gradient of solvent A (5 mM octylamine in water with 5 mM acetic acid) and solvent B (5 mM octylamine in methanol with 5 mM acetic acid), and post-column solvent with 90:10 acetone: DMSO. The gradient was at 0.3 ml min−1: 0 min, 1% B; 3.5 min, 1% B; 4 min, 35% B; 15 min, 35% B; 20 min, 100% B; at 0.4 ml min−1: 20.1 min, 100% B; and 22 min, 100% B, 22.1 min, 1% B; and 27 min, 1% B. Other LC parameters were as follows: post-column flow rate 0.3 ml min−1, column temperature 40 °C and injection volume 5 μl. Other MS parameters were as follows: gas temperature 250 °C, gas flow 9 l min−1, nebulizer pressure 35 psi, sheath gas temperature 250 °C, sheath gas flow 12 l min−1, Vcap 3,500 V and fragmentor 125 V.
Targeted data analysis was performed using both Skyline and MassHunter Profinder software v.10.0 (Agilent Technologies). Further analysis was done using MetaboAnalyst (https://www.metaboanalyst.ca/MetaboAnalyst/ModuleView.xhtml).
D-2HG and L-2HG extraction and analysis
Metabolites were extracted with ice-cold 40:40:20 acetonitrile:methanol:water containing 5 μM L- or D-2-hydroxyglutaric acid-d3 disodium salt (Toronto Research Chemicals, H942578) as an internal standard. After overnight incubation at −80 °C, organoid extract was collected, sonicated and centrifuged at 20,000g for 20 min at 4 °C to precipitate protein. Extracts were then dried in an evaporator (Genevac EZ-2 Elite. For LC–MS analysis, dried samples were derivatized with 100 μl freshly prepared 50 mg ml−1 (+)-diacetyl-l-tartaric anhydride (DATAN; Sigma) in dichloromethane acetic acid (v/v = 4:1) at 75 °C for 30 min. After cooling to room temperature, derivatized samples were dried under nitrogen at room temperature and resuspended in 100 μl UltraPure water (18.2 MΩ, PureLab) before LC–MS/MS analysis. LC–MS analysis was performed on a Thermo Vantage triple-quadrupole mass spectrometer operating in selected reaction monitoring and negative ionization modes using an Acquity UPLC HSS T3 analytical column (2.1 × 100 mm, 1.8 μm, Waters) with an Agilent 1260 infinity binary pump, and applying a gradient of mobile phase A (125 mg l−1 ammonium formate in water adjusted to pH 3.5 with formic acid) and mobile phase B (100% methanol) at a flow rate of 300 μl min−1. The analytical gradient was 0–5 min, 3% B; 5.5–8 min, 80% B. The column was then re-equilibrated for 10 min to ensure retention-time stability. Other LC parameters were as follows: flow rate 300 μl min−1, column temperature 40 °C, sample storage temperature 4 °C and injection volume 10 μl. MS source parameters were as follows: spray voltage 2,500 V, capillary temperature 300 °C, vaporizer temperature 250 °C, sheath gas pressure 50 psi and auxiliary gas pressure 40 psi. Compound-specific S-lens values were as follows: 37 V (2HG) and 41 V (deuterated 2HG). Individual reactions were monitored, and collision energies (CEs) were as follows: 2HG m/z 363.0–147.1 (CE: 12 V), 129.1 (CE: 27 V); deuterated 2HG m/z 368.0–152.1 (CE: 13 V), 132.9 (CE: 22 V). The identities of metabolite enantiomers were determined by comparing with the retention times of the derivatized pure standards. Chromatograms were acquired and processed with TraceFinder software (Thermo Fisher Scientific).
Mitochondria stress test and substrate oxidation assay (Seahorse assays) in organoids
Crypt extraction and organoid preparation
Crypts were extracted as described above. Freshly isolated crypts were resuspended in 1.5 ml Matrigel and kept on ice to prevent the Matrigel from polymerizing during the plating process. The number of organoids to plate was first optimized by testing 1 μl, 2 μl or 3 μl of Matrigel with organoids, and thereafter 2 µl was plated. Samples were plated in the centre of a 96-well Seahorse plate, positioned between the three dots. To ensure consistent conditions, in control wells, the same volumes of Matrigel were plated. To account for variations in plating the same number of organoids in each well, an entire column per condition was plated to have eight replicates. Appropriate organoid culture medium was added to each well, and the plate was kept in the incubator until the day of the experiment. Typically, the organoids were allowed to adapt for two days before the experiment was performed.
Seahorse experiment
The assay was performed as previously described with some adaptations65,66. In brief, the day before the experiment, we prepared complete Seahorse medium: Agilent Seahorse XF (pH 7.4) without phenol red and supplemented with glucose (10 mM), pyruvate (1 mM) and glutamine (2 mM). All reagents were adjusted to a pH of 7.4. The sensor plate was activated in water overnight at 37 °C. On the day of the experiment, the organoids were prepared by removing the organoid complete medium and washing twice with complete Seahorse medium. The complete Seahorse medium was added to the organoids, and they were allowed to adapt for 30 min (maximum one hour) at 37 °C in a non-CO2 incubator.
Injections and settings
For both MitoStress assays (103015-100) and substrate oxidation assays, the following concentrations were injected into the ports: oligomycin (port A) at 50 μM, FCCP at 20 μM and rotenone and antimycin A at 20 μM each. The settings of the XF Analyzer for the assay were as follows: basal (three cycles) with a mix time of 4 min, a wait time of 0 min and a measure time of 3 min; oligomycin (six cycles) with a mix time of 4 min, a wait time of 0 min and a measure time of 3 min; FCCP (three cycles) with a mix time of 4 min, a wait time of 0 min and a measure time of 3 min; and rotenone and antimycin A (three cycles) with a mix time of 4 min, a wait time of 0 min and a measure time of 3 min. For the long-chain fatty acid oxidation stress test (103672-100), we used etomoxir at a concentration of 20 μM supplemented with 500 μM of carnitine; for the glucose/pyruvate oxidation stress test (103673-100) we used UK5099 inhibitor at 4 µM; and for the glutamine oxidation stress test (103674-100) we used CB839 inhibitor at 3 µM.
Normalization
A 4× bright-field picture was taken from all wells to check organoid positioning after the assay. For normalization, we counted the number of organoids per well that were located between the three dots. We found that this normalization worked well for conditions with similar organoid size and viability. For substrate oxidation assays in which organoid samples from the same lineage were treated with different inhibitors, samples were instead normalized by percentage of OCR to correct for differences in organoid number per well, a normalization directly provided by Agilent software (https://seahorseanalytics.agilent.com).
Preparation of single-cell suspensions from intestinal and colonic mucosa for cell sorting and FACS-based immunophenotyping
Intestinal preparation
Crypts were isolated as described above. After spinning, crypts were incubated in DMEM/F-12 medium (D8437, Sigma-Aldrich) supplemented with 0.8 U ml−1 dispase (17105041, Life Technologies, Thermo Fisher Scientific) and 1 mg ml−1 DNase (04716728001, Roche). Cells were incubated in 2 ml of ‘digestion solution’ for 10–15 min at 37 °C, vortexing the samples every 2 min for 30 s. After 10 min, a 20-µl sample was taken and observed under the microscope to check single-cell dissociation. Digestion was stopped by adding 10 ml of fetal bovine serum. Cells were then filtered through a 70-μm mesh, spun down at 290g for 5 min and resuspended in MACS buffer (0.5% BSA and 2 mM EDTA in Ca2+/Mg2+-free PBS). GFPhigh (ISCs), GFPlow (TA cells), GFP− high side scattering and forward scattering (Paneth cells) and the villus fraction (first fraction during mechanical cell extraction) were isolated from Lgr5-EGFP-IRES-creERT2 mice and sorted using a Sony MA900 cell sorter.
Colon preparation
For immunophenotyping of the lamina propria and the muscle layer in the colon of control and DSS-treated mice, samples were prepared as previously reported67. In brief, the entire colon was dissected, opened longitudinally and washed in 1× HBSS (Gibco, 14025-092). The colon was chopped into pieces of 0.5–1 cm and crypts were mechanically dissociated using 8 mM EDTA. The remaining pieces of colon were transferred to digestion mix with dispase and DNAse. Immune cells were further sedimented by centrifugation and immunophenotyping analysis was performed as described below.
FACS-based immunophenotyping
For multiparametric flow-cytometry analysis, cell suspensions were stained with LIVE/DEAD fixable viability dye (1: 500, Invitrogen, R37601) for 30 min in PBS at 4 °C. After this, cells were washed, incubated with Fc block (1: 200, BD Biosciences, 564219) in FACS buffer for 15 min at 4 °C and then stained with a cocktail of conjugated antibodies (see below) for 30 min on ice. After staining, cells were washed three times with FACS buffer and fixed using BD Cytofix/Cytoperm (Thermo Fisher Scientific, 544772) for 20 min at 4 °C, washed again and stored for analysis. Samples were analysed in a BD LSR Fortessa with five lasers, and gates were set by fluorescence-minus-one controls.
The following antibodies were used for flow-cytometry analysis: AF700 CD45 (BioLegend, 103128, clone 30-F11, 1:200), BUV395 CD11b (BD Biosciences, 563553, clone M1/70, 1:200), PE F4/80 (BioLegend, 123110, clone BM8, 1:200), BV605 Ly6G (BD Bioscience, 563005, clone 1A8, 1:200), APC Cy7 Ly6c (BioLegend, 128026, clone HK1.4, 1;200), APC MHCII (BioLegend, 107614, clone M5/114.15.2, 1:200), BV710 CD206 (BioLegend, 141727, clone C068C2, 1:200) and BV650 CD86 (BioLegend, 105035, clone GL-1).
Faecal LCN2 content
Faecal samples were collected longitudinally and analysed for LCN2 levels using an ELISA according to the manufacturer’s instructions (Abcam, ab199083). Frozen faecal samples were reconstituted in PBS containing 0.1% Tween 20 at a concentration of 100 mg ml−1. The samples were vortexed for 20 min to achieve a homogeneous suspension and then centrifuged at 12,000 rpm for 10 min at 4 °C. The clear supernatants were collected and stored at −20 °C until analysis. Measurements were normalized to the weight of the faecal samples.
Immunofluorescence
Mouse tissues
Mouse tissues were fixed overnight at 4 °C in 10% formalin before paraffin embedding. Five-micrometre sections were deparaffinized and rehydrated with Histo-Clear (Thermo Fisher Scientific, National Diagnostics) and an alcohol series and subjected to antigen retrieval by boiling in citrate antigen retrieval buffer (Vector). Slides were blocked in PBS with 5% BSA, and primary antibody staining was performed in blocking buffer + 0.02% Triton X-100 overnight at 4 °C. The following primary antibodies were used: chicken anti-GFP (1:500, Abcam 13970), mouse anti-Ki67 (1:500, BD, 550609), rabbit anti-p53 (1:500, NCL-L-p53-CM5p, Leica Biosystems), rabbit anti-5hmC (1:500, Active Motif, 39769), mouse anti-β-catenin (1:200, BD, 610153), rabbit anti-OGDH (1:100, Proteintech, 15212-1-AP), rabbit anti-VDAC (1:100, Abcam, ab15895), goat anti-ACE2 (1:100, Thermo Fisher Scientific, PA5-47488), rabbit anti-lysozyme (1:500, Thermo Fisher Scientific, MA5-32154), rabbit anti-BrdU (1:100, Abcam, ab6326), rabbit anti-cleaved caspase 3 (1:200, Cell Signaling, 9664S), mouse anti-HNF4α (1:100, Thermo Fisher Scientific, MA1-199), mouse anti-TET1 (1:100, Thermo Fisher Scientific, MA5-16312), rabbit anti-TET2 (1:100, Thermo Fisher Scientific, PA5-85488), rabbit anti-TET3, (1:100, Thermo Fisher Scientific, PA5-31860), rat anti-CD8 (1:200, 14-0808-82, Thermo Fisher Scientific) and rat anti-CD4 (1:100, Thermo Fisher Scientific, 14-9766-82). Primary antibodies were detected with the following fluorescently conjugated secondary antibodies: goat anti-chicken AF488 (Life Technologies A-11039, 1:1,000), goat anti-rabbit AF488 (Life Technologies A-32723, 1:1,000), goat anti-rabbit AF594 (Life Technologies A-11037, 1:1,000), goat anti-mouse AF488 (Life Technologies, A-32723, 1:1,000), goat anti-mouse AF594 (Life Technologies, A-11032, 1:1,000), goat anti-rat AF488 (Life Technologies, A-11006, 1:1,000) and goat anti-rat 594 (Life Technologies, A-11007, 1:1,000). All secondary antibodies were diluted in blocking buffer + 0.02% Triton X-100 and incubated for one hour at room temperature. Slides were then washed with PBS and nuclei were counterstained with PBS containing DAPI and mounted under coverslips with ProLong Gold (Life Technologies).
Human samples
Samples used in human studies were obtained from TissueArray (https://www.tissuearray.com/). Specifically, tissue microarrays (TMAs) CO809b, CO246, CO245a were used. Immunofluorescence staining was performed as described above, using the same primary and secondary antibodies.
Organoid immunofluorescence
Organoids were recovered using Cell Recovery Solution (354253, Corning BD) and fixed with 4% paraformaldehyde (PFA) for 20 min at room temperature (18–21 °C). Next, the samples were passed through an ethanol series (70%, 96% and 100%) and embedded in paraffin. Immunohistochemistry and immunofluorescence were performed using standard techniques as described above, using the same primary and secondary antibodies.
Image acquisition and analysis
Images were acquired with a Zeiss AxioImager microscope using Axiovision software. Five to ten images per slide were obtained. Quantification was performed either by counting the number of positive cells per crypt or villus (at least 50 crypts and 100 villi were quantified) or by calculating the percentage of the positive area using the Color Deconvolution plug-in in ImageJ v.1.7 software. Additional macros were developed by the authors to quantify immunofluorescence images.
Multiplex immunofluorescence in human and mouse FFPE tissues
Multiplex immunofluorescence imaging was performed using the Comet Lunaphore platform68 for mouse tissues and the CellDive platform69 for human TMAs of formalin-fixed paraffin-embedded (FFPE) tissues. Tissue processing was done as described above (‘Immunofluorescence’ section), with the key difference that antigen retrieval included the use of two buffers. Specifically, after deparaffinization, slides were boiled in citrate antigen retrieval buffer (pH 6.0, Vector Labs H-3300-250) and then directly transferred to Tris-EDTA unmasking solution (pH 9.0, Vector Labs H-3301-250) and boiled again. This dual antigen retrieval method improved antigen unmasking in our samples, enabling the combination of antibodies that work with either citrate or EDTA buffers. After antigen retrieval, the slides were loaded onto the respective platforms. For details on the antibody panels and staining conditions, see Supplementary Tables 4 (COMET Lunaphore mouse panel) and 5 (CellDive human panel). For CellDive multiplex immunofluorescence, the first round of staining was performed using primary and secondary antibodies. In most cases, subsequent rounds used primary conjugated antibodies. In addition, a stripping step was performed as previously described70, allowing for a second round of primary and secondary antibody staining on the same TMAs. The HNF4α antibody was self-conjugated according to the manufacturer’s instructions (https://www.thermofisher.com/order/catalog/product/A20186).
smFISH
smFISH analysis was performed as previously described, with slight modifications in the protocol71.
Probe design for multiplex smFISH
We built on published software72,73 to design custom panels for smFISH. This design strategy relies on the precomputation of all possible 30-mer sequences found in mouse cDNAs (Ensembl GRCm38.p6), augmented with coding sequences of fluorescent proteins engineered into our mouse model. We excluded pseudogenes from the potential pool of mRNAs to design probes for. We computed multiple scores for each 30-mer, including Tm, GC content and potential for hybridization with ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs). We used the following criteria for including a 30-mer in our candidate probe set: GC content 43–63% and Tm (66–76 °C), excluding 30-mers that contain at least a 15-mer present in an rRNA or a tRNA. In addition, we computed expression-informed penalties to estimate the specificity of each candidate probe. We adapted published software72,73 to include single-cell information into the estimation of specificity scores. We reasoned that incorporating single-cell information would decrease the chances of selecting probes with off-target binding to highly expressed genes in rare cell populations. To do so, we used the published single-cell data used in figure 1 in ref. 26. Following suggested parameters from the original MERFISH publications72,73, we considered 30-mers with a specificity score greater than 0.75 as candidates for our panels. We aimed to select 92 non-overlapping probes per gene. Whenever this was not possible owing to transcript length, homology to other genes or other sequence properties, we allowed a maximum overlap of 20 bp between probes. Probe sequences can be found in Supplementary Table 3.
Sample preparation
‘Swiss-rolled’ intestines were positioned in a cassette and fixed with 4% PFA 1× PBS solution for four hours at 4 °C. Cassettes were then transferred to 4% PFA and 30% sucrose in 1× PBS and incubated overnight at 4 °C. To preserve villus structures, cassettes were transferred for four hours to 30% sucrose and 50% OCT for three hours at room temperature and finally embedded in Tissue Plus OCT Compound (Fisher Healthcare, 4585) in a cryomold (Tissue-Tek, 4557). Moulds were placed on dry ice until all OCT was frozen, and intestinal samples were stored at −80 °C.
Coverslip preparation
Coverslips for smFISH staining were prepared as previously described71. In brief, 40-mm-diameter coverslips (Bioptechs, 0420-0323-2) were cleaned by immersing them in a 1:1 mix of 37% HCl and methanol at room temperature for 30 min. Coverslips were then washed with Milli-Q water, washed once with 70% ethanol and then gently dried with nitrogen gas. Cleaned coverslips were submerged in 0.1% (v/v) triethylamine (Millipore, TX1200) and 0.2% (v/v) allyltrichlorosilane (Sigma, 107778) in chloroform for 30 min at room temperature. They were washed once with chloroform and once with 100% ethanol, then dried using nitrogen gas. Coverslips were stored long term in a desiccated chamber at room temperature.
Poly-lysine coating of coverslips
To prepare coverslips for staining individual samples, pre-treated coverslips were coated with 0.1 mg ml−1 poly-d-lysine (Thermo Fisher Scientific, A3890401) for one hour at room temperature. Next, they were washed once with 1× PBS, and three times with nuclease-free water. After that, they were left to dry for at least two hours before sectioning the tissue.
Tissue sectioning, fixation and permeabilization for smFISH staining
Tissue sections of 10-μm thickness were mounted into poly-d-lysine-coated coverslips. Coverslips were dried for 5–10 min at 50 °C and placed on dry ice until completion of sectioning of all samples. Next, plates with coverslips were transferred to ice, and treated with 3 ml 1× PBS, followed by fixation at room temperature with 4% PFA 1× PBS for 10 min. Coverslips were then washed three times with 1× PBS and maintained at 4 °C overnight in ice-cold 70% ethanol for permeabilization.
Pre-staining treatment of permeabilized tissues
After overnight incubation, coverslips were rehydrated with 1× PBS on ice for 10 min. To bleach endogenous fluorescence of lineage reporters and reduced autofluorescence from lysozyme granules, tissues were incubated with 3% hydrogen peroxide (Thermo Fisher Scientific, H325-500), 1:600 37% HCl (v/v) 1× PBS and placed under a heat lamp for one hour at room temperature. They were then washed twice with 1× PBS and once with 2× SSC. Next, they were treated with digestion solution (pre-warmed at 37 °C) containing a final concentration of 20 μg ml−1 proteinase K (Sigma, 3115836001) in 2× SSC solution, and incubated at 37 °C for 10 min. Next, coverslips were washed three times with 2× SSC and treated with pre-hybridization solution (30% formamide (Thermo Fisher Scientific, AM9344) and 2× SSC) and incubated for at least three hours at 37 °C.
Staining with primary probes
Primary probes were diluted to 100 nM per probe in 3H staining buffer (30% formamide, 10% dextran sulfate (Sigma-Aldrich, D8906-50G), 1 mg ml−1 yeast tRNA (Thermo Fisher Scientific, 15401029) and 2× SSC). In addition, 2 μM anchor probe was added to the staining solution containing specific probes. A 100-µl droplet of this solution was then added to coverslips after their pre-hybridization incubation, then the coverslips were placed on a 15-cm dish with a wet Kimwipe used as a humidity buffer and incubated at 37 °C for 36–48 h. Next, post-hybridization wash buffer (30% formamide and 2× SSC) was pre-heated to 37 °C. Coverslips were washed twice with post-hybridization wash buffer at 47 °C for 30 min. Finally, coverslips were transferred to 2× SSC solution and maintained at 4 °C until the next step.
Gel embedding and digestion
Samples were embedded on a thin layer of polyacrylamide gel, to allow subsequent tissue clearing through digestion of protein and lipids. The gel solution was composed of 4% (v/v) 19:1 acrylamide/bis-acrylamide (Bio-Rad, 1610144), 60 mM Tris⋅HCl pH 8 (Invitrogen, 15568-025) and 0.3 M NaCl (Boston BioProducts, R-244), supplemented with the polymerizing agents ammonium persulfate (Sigma, 09913) and TEMED (Sigma, T7024) at final concentrations of 0.03% (w/v) and 0.15% (v/v), respectively, and polymerized as previously described71. Polymerization was complete after two hours at room temperature. Next, gel-embedded coverslips were transferred to a 6-cm tissue culture dish with 2× SSC. Gel-embedded samples were treated overnight at 37 °C with digestion solution: 2% SDS (Invitrogen, AM9822), 0.25% Triton X-100 (Acros Organics, 327371000) and a 1:100 dilution of proteinase K (NEB, P8107S) in 2× SSC. After overnight digestion, samples were washed for 30 min with 2× SSC and gentle agitation.
Staining with secondary probes
We used readout probes consisting of a 20-bp oligonucleotide conjugated to a fluorophore (Alexa Fluor 488, Cy3B, Cy5 or Alexa Fluor 750) through a disulfide bond. Fluorescent conjugated probes were purchased from Bio-Synthesis. The secondary staining solution was composed of 5% ethylene carbonate (Sigma-Aldrich, E26258-100G) in 2× SSC. The secondary staining solution was supplemented by a secondary readout probe for each fluorescent colour at a final concentration of 3 nM, and with DAPI at a final concentration of 1 μM. Secondary staining was performed following the same procedure as the primary staining step, with the exception that it was done for 20 min at room temperature, covering samples with aluminium foil. After hybridization, samples were washed once with 10% ethylene carbonate in 2× SSC for 20 min with gentle agitation, and three times with 2× SSC for 5 min per wash.
Iterative smFISH imaging
Iterative smFISH imaging was performed as previously described71. Combinations of readout sequences and target mRNA species are provided in Supplementary Table 3.
smFISH image processing and analysis
To collapse z-stacks into a single two-dimensional image, maximum projection images were generated using the Nikon Elements software’s maximum projection function. After each round of FISH imaging, we took an additional image with cleaved fluorophores to capture the background signal for each channel. Because the microscope has unequal sensitivity to the five fluorophores, we also imaged each fluorophore’s flat field to capture its bias. We corrected raw FISH images by subtracting the background signal of each gene and then dividing by the flat-field bias of the conjugated fluorophore, then thresholding to 0 to correct any negative-valued pixels. Additional alignment was performed using DAPI.
Transmission electron microscopy
Duodenal samples from C57Bl/6 mice were collected and prepared as described above. One- to two-millimetre intestinal samples were fixed in 2% glutaraldehyde, 4% PFA and 2 mM CaCl2 in 0.1 M sodium cacodylate buffer, pH 7.2, at room temperature for more than one hour, dehydrated in an acetone series, post-fixed in 1% osmium tetroxide and processed for Eponate 12 (Ted Pella) embedding. Ultrathin sections (65 nm) were cut, post-stained with uranyl acetate and lead citrate and imaged in a Tecnai 12 electron microscope (FEI), operating at 120 kV and equipped with an AMT BioSprint29 digital camera (AMT Imaging). Mitochondrial quantification was performed as previously described74.
Immunoblotting
Immunoblotting was performed in mouse ES cells containing a doxycycline-inducible GFP-coupled shRNA30,33. The genotypes used were shOgdh_2081, shOgdh_346 (ref. 1) and shRen_731. Mouse ES cells were treated with doxycycline for 72 h. In brief, the supernatant was removed, and the cells were washed three times with PBS. Mouse ES cells were then lysed using RIPA lysis buffer (Sigma-Aldrich, R0278) supplemented with NaF (1 mM), Na4P2O7 (20 mM) and Na3VO4 (2 mM). The lysates were incubated for 15 min on ice and subsequently clarified by centrifugation at 4 °C and 10,000g. The protein concentration was determined using the Pierce BCA protein assay kit (23225, Thermo Fisher Scientific) following the manufacturer’s instructions. Lysates were concentrated to a final concentration of 1 mg ml−1 by boiling the appropriate amount of protein lysate with 2× Laemmli buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol and 0.004% bromophenol blue in 0.2 M Tris-HCl, pH 7) at 90 °C for 10 min. Twenty micrograms of protein lysates were loaded onto SDS–PAGE gels and transferred to 0.2-μm nitrocellulose membranes (LI-COR Biosciences, 926-31090). The membranes were blocked with 5% blotting-grade blocker (non-fat dry milk, 170-6404, Bio-Rad) in Tris-buffered saline containing 1% Tween 20 (TBS-T) for one hour at room temperature. Then the membranes were incubated overnight at 4 °C in TBS-T with 3% sodium azide with rabbit anti-OGDH antibody (1:500, Proteintech, 15212-1-AP). After washing the membranes three times with TBS-T for 10 min each at room temperature, they were incubated with secondary anti-rabbit antibody (1:5,000, Cell Signaling, 7074S) in 1% blotting-grade blocker. The protein bands were visualized using enhanced chemiluminescence (ECL) detection reagent (Cell Signaling, 6883P3) according to the manufacturer’s instructions.
ChIP in intestinal crypts
Intestinal crypts were isolated and cross-linked for 10 min at room temperature in 1% formaldehyde. Cross-linking reactions were stopped by adding 1.25 M glycine to a final concentration of 125 mM. The crypts were then centrifuged for 10 min at 4 °C and washed in cold PBS. The cells were lysed with 1 ml lysis buffer 1 (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100 and 1× protease inhibitor) followed by centrifugation at 4 °C. The pellet was resuspended in lysis buffer 2 (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA and 1× protease inhibitors). Finally, the pellets were resuspended in 1 ml lysis buffer 3 (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM N-lauroylsarcosine and 1× protease inhibitors) and 100 μl 10% Triton X-100 was added. The samples were sonicated for 10 min using an E220 Focused Ultrasonicator (Covaris, PN 500239). Samples were sonicated using milliTube 1 ml with AFA fibre (Covaris, PN 520130) under flowing conditions (140 pip, 5% duty cycle, 200 CBP). The soluble fraction was quantified using the Bradford assay, and 400 μg of the soluble fraction was used for immunoprecipitation of the transcription factors HNF4α (1 μg, Thermo Fisher Scientific, MA1-199), and SMAD4 (1 μg, Cell Signaling, 46535) with rabbit IgG (1 μg, Cell Signaling, 2729S) and mouse IgG (1 μg, Santa Cruz Biotechnology, sc-2025) used as a control. The chromatin and antibody mixtures were incubated overnight at 4 °C in a total volume of 500 μl. The immunoprecipitated mixture was then washed sequentially with a Triton dilution buffer (1% Triton X-100, 2 mM EDTA pH 8, 150 mM NaCl and 20 mM Tris-HCl pH 8.1), mixed micelle wash buffer (1% Triton X-100, 5 mM EDTA pH 8, 150 mM NaCl, 20 mM Tris-HCl pH 8.1, 5% sucrose, 0.2% NaN3 and 0.2% SDS), buffer 500 (0.1% w/v deoxycholic acid, 1 mM EDTA pH 8, 50 mM HEPES pH 7.5, 500 mM NaCl, 1% Triton X-100, 0.2% NaN3) and a LiCl wash buffer (0.5% w/v deoxycholic acid, 1 mM EDTA pH 8, 250 mM LiCl, 0.5% v/v NP-40, 10 mM Tris-HCl and 0.2% NaN3). The samples were de-cross-linked, and the DNA was extracted using phenol, chloroform and isoamyl alcohol mixtures, washed with 80% ethanol and resuspended in 200 μl TE buffer. qRT–PCR was performed using specific primers (Supplementary Table 2).
Statistical analysis and data representation
Before performing any statistical test, we tested for normal distribution using the D’Agostino–Pearson test. For continuous variables, we used the t-test, Mann–Whitney U test, one‐way ANOVA or Friedman’s test. For categorical variables, we used the chi‐squared test or Fisher’s exact test. The Mantel–Cox test was used to analyse the Kaplan–Meier survival of mice. If significant differences by one-way ANOVA were found, group-wise comparisons were done using Tukey’s multiple comparisons test. If significant differences by Friedman’s test were found, Dunn’s multiple comparisons test was used. The predictive value of Ogdh expression in enterocytes, ISCs and Paneth cells was evaluated by examining the area under the receiver operating characteristic (ROC) curve with a confidence interval of 95%. All statistical tests were considered statistically significant when P was less than 0.05. Statistical significance in figures is summarized as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ****P ≤ 0.0001 between the means of a minimum of three samples. Results are expressed as mean ± s.e.m.
The immunofluorescence, H&E, other stainings and IHC data shown are representative of at least three independent mice. Quantification of immunofluorescence, H&E and other stainings was performed for more than 50 crypts or 100 villi per mouse in at least 3 independent mice. For in vitro experiments, at least three independent experiments were performed. For the in vivo experiments, at least five mice per group were used.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.