Protein sequences
Human cDNAs were cloned from RPE-1 and U2OS cells by total RNA isolation, oligo-dT reverse transcription, followed by PCR with gene-specific oligonucleotides and cloning into expression vectors. Sanger sequencing showed that the cloned cDNA coded for the reference protein sequence for the following proteins: ORC1, ORC2, ORC3 (NCBI: iso2, Uniprot: canonical iso), ORC4, ORC5, ORC6, MCM2, MCM3, MCM5, MCM6, MCM7, CDC6 and GMNN. For MCM4 and CDT1, the cloned cDNA coded for a natural variant that differed from NCBI reference protein sequences, but according to the Genome Aggregation Database (https://gnomad.broadinstitute.org) represented the majority allele (allele frequency > 0.5): MCM4_L650M (allele frequency: 0.862), CDT1_C234R (allele frequency: 0.999). These alleles were used and considered wild type throughout this study.
Expression and purification of human MCM2â7
MCM2â7 was expressed in insect cells using the biGBac baculovirus expression vector system10. Human cDNAs of MCM2, MCM3, MCM4, MCM5, MCM6 and MCM7 were cloned into pLIB. MCM3 was subcloned to contain an N-terminal TEV protease-cleavable Flag tag. pLIB-derived expression cassettes of MCM4, MCM5, MCM6, MCM7 were subcloned into pBIG1a (pBIG1a:MCM4, MCM5, MCM6, MCM7) and expression cassettes of MCM2 and FlagâMCM3 were subcloned into pBIG1b (pBIG1b:MCM2, FlagâMCM3). Expression cassettes of these two vectors were subcloned into pBIG2ab (pBIG2ab:MCM4, MCM5, MCM6, MCM7, MCM2, FlagâMCM3). Baculovirus was generated and amplified in Sf9 cells (Thermo Fisher, 12659017) using the EMBacY baculoviral genome41. For protein expression, Sf9 cells were infected and collected 52âh after infection, flash-frozen and stored at â80â°C.
Cell pellets were thawed on ice in MCM buffer (50âmM HEPES/KOH pH 7.6, 100âmM potassium glutamate, 5âmM magnesium acetate, 10% glycerol, 0.02% NP-40, 1âmM dithiothreitol (DTT)) + protease inhibitors (1 tablet per 50âml Roche Complete Ultra EDTA-free, 10âµgâmlâ1 leupeptin, 10âµgâmlâ1 pepstatin A, 1âmM AEBSF, 1âµgâmlâ1 aprotinin, 2âmM benzamidine). Cells were resuspended and lysed by Dounce homogenization using a tight-fitting pestle. The lysate was cleared by centrifugation (158,000g, 4â°C, 1âh) and incubated with anti-Flag M2 affinity gel (Sigma-Aldrich, A2220) for 1âh at 4â°C with rotation. The beads were transferred to a Econo-Pac gravity flow column (Bio-Rad, 732-1010) and washed with 2à 20 column volumes MCM buffer, 1à20âcolumn volumes MCM buffer + 5âmM ATP (Sigma-Aldrich, A2383). For dephosphorylation, the beads were resuspended in MCM buffer + 0.2âmgâmlâ1 lambda protein phosphatase, 1âmM manganese (ii) chloride and incubated for 1âh at 4â°C. The beads were washed with 2à 20âcolumn volumes MCM buffer. For elution and proteolytic tag removal the beads were resuspended in 5âcolumn volumes MCM buffer + 80âµgâmlâ1 TEV protease, 0.1âmgâmlâ1 3ÃFlag peptide for 2âh at 4â°C. The eluate was concentrated using an Amicon Ultra-15 concentrator (Merck, UFC903024) and further purified by gel filtration using a HiLoad 16/600 Superdex 200âpg column equilibrated in MCM buffer. Fractions containing stoichiometric MCM2â7 were concentrated, flash-frozen in liquid nitrogen, and stored at â80â°C.
Expression and purification of human ORC1â5
ORC1â5 was expressed in baculovirus-infected insect cells. The coding sequences of human ORC1, ORC2, ORC3, ORC4 and ORC5 were codon-optimized for Spodoptera frugiperda, synthesized (GeneArt, Thermo Fisher Scientific) and subcloned into modified pBIG1 vectors that contain a pLIB-derived polyhedrin expression cassette. ORC1 was subcloned into pBIG1a with a TEV protease-cleavable N-terminal 3à Flag tag. ORC2 was subcloned into pBIG1b, ORC3 into pBIG1c, ORC4 into pBIG1d, ORC5 into pBIG1e. Expression cassettes from these five vectors were subcloned into pBIG2abcde (pBIG2abcde:Flag-ORC1,2,3,4,5). Baculoviruses were generated using EMBacY and Sf9 cells. To express ORC1â5, Sf9 cells were infected, collected 52âh after infection, flash-frozen and stored at â80â°C.
Cell pellets were thawed in ORC1â5 lysis buffer (50âmM HEPES/KOH 7.6, 650âmM potassium chloride, 5âmM magnesium acetate, 1âmM ATP, 10% glycerol, 0.02% NP-40, 1âmM DTT, 2âmM benzamidine) + protease inhibitors (1 tablet per 50âml Roche Complete Ultra EDTA-free, 10âµgâmlâ1 leupeptin, 10âµgâmlâ1 pepstatin A, 1âmM AEBSF, 1âµgâmlâ1 aprotinin). Cells were lysed by Dounce homogenization, and the lysate cleared by centrifugation (158,000g, 4â°C, 1âh). ORC1â5 was bound to anti-Flag M2 affinity gel for 1âh at 4â°C. The column was washed with 2à 20âcolumn volumes ORC1â5 lysis buffer + 4âmM ATP. For dephosphorylation, the beads were resuspended in 1âcolumn volumes ORC1â5 lysis buffer + 0.2âmgâmlâ1 lambda protein phosphatase, 1âmM manganese (ii) chloride, potassium chloride adjusted to 650âmM, and incubated at 4â°C for 1âh. The column was washed with ORC1â5 lysis buffer. For elution and proteolytic tag removal, the beads were resuspended in 5âcolumn volumes ORC1â5 lysis buffer + 80âµgâmlâ1 TEV protease, 0.1âmgâmlâ1 3à Flag peptide, potassium chloride adjusted to 650âmM and were incubated at 4â°C for 2âh. To remove TEV protease, the eluate was supplemented with 35âmM imidazole pH 8.0 and incubated with Ni-NTA Agarose (Invitrogen, R90115) for 1âh at 4â°C. The flowthrough was concentrated and further purified by gel filtration using a HiLoad 16/600 Superdex 200âpg column (Cytiva) equilibrated in ORC1â5 SEC buffer (50âmM HEPES/KOH pH 7.6, 650âmM potassium chloride, 5âmM magnesium acetate, 10% glycerol, 0.02% NP-40, 1âmM DTT). Fractions containing stoichiometric ORC1â5 were concentrated, flash-frozen, and stored at â80â°C.
Expression and purification of human CDC6
CDC6 was expressed in insect cells. Human cDNA of CDC6 was cloned into pLIB to contain an N-terminal TEV protease-cleavable Flag tag. The baculovirus was generated in Sf9 cells using the EMBacY genome. For expression, Sf9 cells were infected and the culture collected 52âh after infection. The cell pellet was flash-frozen in liquid nitrogen and stored at â80â°C.
The cell pellet was thawed on ice in CDC6 lysis buffer (50âmM HEPES/KOH pH 7.6, 650âmM potassium chloride, 5âmM magnesium acetate, 4âmM benzamidine, 1âmM ATP, 10% glycerol, 0.02% NP-40, 1âmM DTT) + protease inhibitors (1 tablet per 50âml Roche Complete Ultra EDTA-free, 10âµgâmlâ1 leupeptin, 10âµgâmlâ1 pepstatin A, 1âmM AEBSF, 1âµgâmlâ1 aprotinin). Cells were lysed with a Dounce homogenizer and the lysates centrifuged (158,000g, 4â°C, 1âh). The cleared lysate was incubated with anti-Flag M2 affinity gel for 1âh at 4â°C. The beads were washed with 2à 20âcolumn volumes CDC6 lysis buffer + 4âmM ATP. For dephosphorylation, the beads were resuspended in CDC6 lysis buffer + 0.2âmgâmlâ1 lambda protein phosphatase, 1âmM manganese (ii) chloride and incubated for 1âh at 4â°C. The column was washed with CDC6 lysis buffer, followed by CDC6 HTP-wash buffer (50âmM potassium phosphate pH 7.6, 75âmM potassium acetate, 5âmM magnesium acetate, 0.1% Triton X-100, 1âmM DTT, 2âmM ATP). For elution, the beads were resuspended in CDC6 HTP-wash buffer + 80âµgâmlâ1 TEV protease, 0.1âmgâmlâ1 3à Flag peptide and incubated for 2âh at 4â°C. To remove TEV protease, the eluate was supplemented with 30âmM imidazole pH 8.0 and incubated with Ni-NTA agarose for 1âh at 4â°C. The CDC6-containing flowthrough was collected. A hydroxyapatite column was prepared by resuspending 2âg Bio-Gel HTP Hydroxyapatite (Bio-Rad, 130-0420) in 12âml CDC6 HTP-wash buffer. To remove fine particles, the beads were allowed to settle for 2âmin and the supernatant was removed. Two more times, the beads were resuspended in 10âml CDC6 HTP-wash buffer, allowed to settle for 2âmin, and the supernatant removed. Then, 4âml of a 50% slurry were incubated with CDC6 for 15âmin at 4â°C. The beads were transferred to a gravity flow column and washed with 2âml CDC6 HTP-wash buffer, followed by 5âml CDC6 HTP-rinse buffer (50âmM potassium phosphate pH 7.6, 150âmM potassium acetate, 5âmM magnesium acetate, 0.1% Triton X-100, 15% glycerol, 1âmM DTT). CDC6 was eluted by applying 10âml CDC6 HTP-elution buffer (50âmM potassium phosphate pH 7.6, 400âmM potassium acetate, 5âmM magnesium acetate, 0.1% Triton X-100, 15% glycerol, 1âmM DTT). The eluate was dialysed 2Ã1âh at 4â°C against CDC6 dialysis buffer (50âmM HEPES/KOH pH 7.6, 650âmM potassium chloride, 5âmM magnesium acetate, 10% glycerol, 0.02% NP-40, 1âmM DTT). CDC6 was concentrated, flash-frozen, and stored at â80â°C.
Expression and purification of human CDT1
CDT1 was expressed in insect cells. The coding sequence of human CDT1 was codon-optimized for S. frugiperda, synthesized (GeneArt, Thermo Fisher Scientific) and subcloned into pLIB as a fusion protein with an N-terminal FlagâHisâSumoEu1 fusion42. The baculovirus was generated in Sf9 cells using the EMBacY genome. Expression cultures were collected 52âh after infection, snap-frozen in liquid nitrogen and stored at â80â°C.
The cell pellet was thawed in CDT1 buffer (50âmM HEPES pH 7.6, 650âmM potassium chloride, 5âmM magnesium acetate, 10% glycerol, 0.02% NP-40, 1âmM DTT) + protease inhibitors (1 tablet per 50âml Roche Complete Ultra EDTA-free, 10âµgâmlâ1 leupeptin, 10âµgâmlâ1 pepstatin A, 1âmM AEBSF, 1âµgâmlâ1 aprotinin). Cells were lysed using a Dounce homogenizer. The lysate was cleared by centrifugation (158,000g, 4â°C, 1âh) and incubated with anti-Flag M2 affinity gel for 1âh at 4â°C. The beads were transferred to a gravity flow column and washed twice with CDT1 buffer + 5âmM ATP. The beads were resuspended in CDT1 buffer + 0.2âmgâmlâ1 lambda protein phosphatase, 1âmM manganese (ii) chloride and incubated for 1âh at 4â°C. The beads were washed with CDT1 buffer. For proteolytic elution the beads were resuspended in CDT1 buffer + 80âµgâmlâ1 His-SENP1_EuH protease42 and incubated for 2âh at 4â°C. To remove His-SENP1_EuH protease, 35âmM imidazole pH 8.0 was added and the eluate incubated with Ni-NTA agarose for 1âh at 4â°C. The CDT1-containing flowthrough was concentrated and further purified by gel filtration on a Superdex 200 Increase 10/300GL column (Cytiva) using CDT1 buffer. CDT1-containing fractions were concentrated, snap-frozen, and stored at â80â°C.
Expression and purification of human ORC6
ORC6 was expressed in Escherichia coli. The coding sequence of human ORC6 was codon-optimized for E. coli, synthesized (GeneArt, Thermo Fisher Scientific) and subcloned into pK27Sumo to encode the fusion protein HisâSumoâORC6. The protein was expressed using the strain T7express lysY (NEB, C3010I). Expression was induced with 0.4âmM IPTG at OD600â~â0.6 at 16â°C. The culture was collected after 16âh, flash-frozen, and stored at â80â°C.
The cell pellet was thawed in ORC6 lysis buffer (50âmM HEPES pH 7.6, 500âmM potassium chloride, 5âmM magnesium acetate, 10% glycerol, 0.02% NP-40, 1âmM DTT, 35âmM imidazole) + protease inhibitors (1 tablet per 50âml Roche Complete EDTA-free, 1âmM AEBSF), lysozyme. The cells were resuspended and lysed by sonication. The lysate was centrifuged (158,000g, 4â°C, 1âh) and the cleared lysate incubated with Ni-NTA agarose (Invitrogen) for 1âh at 4â°C. The beads were transferred to a gravity flow column and washed twice with ORC6 lysis buffer + 5âmM ATP and twice with ORC6 lysis buffer. The protein was eluted with ORC6 elution buffer (50âmM HEPES/KOH pH 7.6, 200âmM potassium chloride, 5âmM magnesium acetate, 10% glycerol, 0.02% NP-40, 1âmM DTT, 300âmM imidazole pH 8.0). The Sumo-specific protease His-Ulp1 was added at 0.08âmgâmlâ1 and the protein was dialysed overnight against ORC6 SEC buffer (50âmM HEPES/KOH pH 7.6, 200âmM potassium chloride, 5âmM magnesium acetate, 10% glycerol, 0.02% NP-40, 1âmM DTT). To remove His-Ulp1 protease and HisâSumo, the imidazole concentration was adjusted to 35âmM and the protein incubated with Ni-NTA agarose (Invitrogen) for 1âh at 4â°C. The ORC6 containing flowthrough was concentrated and further purified by gel filtration using a HiLoad 16/600 Superdex 200âpg column (Cytiva) equilibrated in ORC6 SEC buffer. ORC6 containing fractions were pooled, snap-frozen, and stored at â80â°C.
Expression and purification of truncated loading factors
The ORC1â5 complex with ORC1(âN) (ORC1â5(1âN)), and CDC6(âN) and CDT1(âN) with N-terminal IDR truncations were expressed in insect cells using a FlagâHisâSumoEu1 fusion system42 and sequences that were codon-optimized for S. frugiperda and synthesized (GeneArt, Thermo Fisher Scientific). The SumoEu1 fusions were only partially stable in Sf9 cells giving relatively low yields. FlagâHisâSumoEu1âORC1(391â861) was cloned into pBIG1a containing a pLIB-derived polyhedrin expression cassette. The expression cassette was subcloned together with ORC2â5 expression cassettes into pBIG2abcde (pBIG2abcde:FlagâHisâSumoEu1âORC1(âN), ORC2â5). FlagâHisâSumoEu1âCDC6(âN) (CDC6 residues 143â560) and FlagâHisâSumoEu1âCDT1(âN) (CDT1 residues 167â546) were cloned into pLIB. Baculoviruses were generated in Sf9 cells using EMBacY. Sf9 expression cultures were collected 52âh after infection, flash-frozen and stored at â80â°C.
The three proteins were purified with the same purification protocol with only the gel filtration column differing. Cell pellets were thawed in Wash-300 buffer (50âmM HEPES/KOH pH 7.6, 300âmM potassium chloride, 5âmM magnesium acetate, 1âmM ATP, 10% glycerol, 0.02% NP-40, 1âmM DTT) + protease inhibitors (1 tablet per 50âml Roche Complete Ultra EDTA-free, 10âµgâmlâ1 leupeptin, 10âµgâmlâ1 pepstatin A, 1âmM AEBSF, 1âµgâmlâ1 aprotinin, 4âmM benzamidine) and lysed using a Dounce homogenizer. The lysate was centrifuged (158,000g, 4â°C, 1âh) and the cleared lysate incubated with anti-Flag M2 affinity gel for 2âh at 4â°C. The beads were washed twice with Wash-300 buffer + 4âmM ATP, and then resuspended in Wash-300 buffer + 0.2âmgâmlâ1 lambda protein phosphatase, 1âmM manganese (ii) chloride and incubated for 1âh at 4â°C. The beads were washed with Wash-300 buffer. For proteolytic elution, the beads were resuspended in Wash-300 buffer + 80âµgâmlâ1 His-SENP1_EuH protease42 and incubated for 2âh at 4â°C. To remove His-SENP1_EuH protease, the eluate was supplemented with 35âmM imidazole pH 8.0 and incubated with Ni-NTA agarose (Invitrogen) for 1âh at 4â°C. The flowthrough was concentrated and further purified by gel filtration using SEC-âN buffer (50âmM HEPES/KOH pH 7.6, 500âmM potassium glutamate, 5âmM magnesium acetate, 10% glycerol, 0.02% NP-40, 1âmM DTT). ORC1â5(1âN) was purified using a HiLoad 16/600 Superdex 200âpg column (Cytiva). CDC6(âN) and CDT1(âN) were purified using a Superdex 200 Increase 10/300GL column (Cytiva). Fractions containing the respective protein were concentrated, snap-frozen, and stored at â80â°C.
Expression and purification of human geminin
Geminin was expressed in insect cells. Human cDNA of geminin was cloned into pLIB with an N-terminal Flag tag. The baculovirus was generated in Sf9 cells using EMBacY. Geminin was expressed in Sf9 cells and the culture collected 52âh after infection. The cell pellet was stored at â80â°C.
The cell pellet was thawed in GMNN lysis buffer (50âmM HEPES/KOH pH 7.6, 300âmM potassium chloride, 5âmM magnesium acetate, 10% glycerol, 0.02% NP-40, 1âmM DTT) + protease inhibitors (1 tablet per 50âml Roche Complete Ultra EDTA-free, 10âµgâmlâ1 leupeptin, 10âµgâmlâ1 pepstatin A, 1âmM AEBSF, 1âµgâmlâ1 aprotinin) and lysed using a Dounce homogenizer. The lysate was cleared by centrifugation (158,000g, 60âmin, 4â°C) and incubated with anti-Flag M2 affinity gel for 1âh. The beads were washed with GMNN lysis buffer + 5âmM ATP, followed by GMNN lysis buffer. Protein was eluted in GMNN lysis buffer + 0.1âmgâmlâ1 3ÃFlag peptide. The protein was concentrated and further purified by gel filtration on a Superdex 200 Increase 10/300GL column equilibrated in GMNN SEC buffer (50âmM HEPES/KOH pH 7.6, 200âmM potassium chloride, 5âmM magnesium acetate, 10% glycerol, 0.02% NP-40, 1âmM DTT). Geminin containing fractions were concentrated and the protein concentration determined for the homodimer. The protein was flash-frozen, and stored at â80â°C.
Mass spectrometry
Protein preparations of human MCM2â7, ORC1â5 (FL), ORC1â5(1âN), CDC6 (FL), CDC6(âN), CDT1 (FL) and CDT1(âN) were subjected to mass spectrometry. Insect cell ORC6 and CDC6 were not detected in any purification.
Nuclease footprinting assay with human proteins
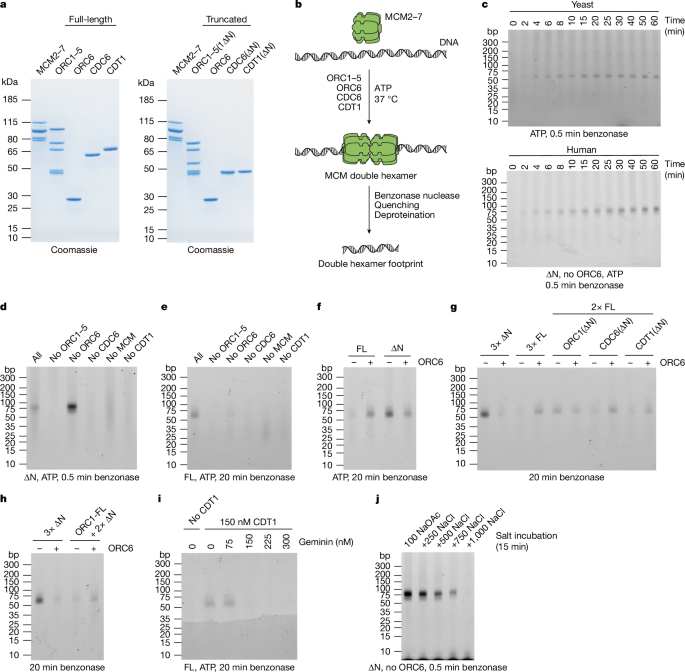
MCM loading reactions were performed in assay buffer (25âmM HEPES/KOH pH 7.6, 100âmM sodium acetate, 10âmM magnesium acetate, 1âmM DTT). MCM was typically loaded onto a ARS1-containing 10.6âkb plasmid that was purified by caesium chloride density gradient centrifugation (pJY2243).
A 20âµl reaction with truncated proteins typically contained 4ânM (27.5ângâµlâ1) plasmid DNA (10.6âkb), 2âmM ATP, 60ânM MCM2â7, 150ânM ORC6, 150ânM CDC6(âN), 150ânM CDT1(âN), and 90ânM ORC1â5(1âN). Stocks of MCM2â7, CDC6(âN), CDT1(âN), and ORC1â5(1âN) were diluted to a 10à working concentration in assay buffer. ORC6 was diluted to a 20à working concentration in assay buffer + 200âmM sodium chloride. Reactions were started by adding MCM2â7, ORC6, CDC6(âN), CDT1(âN) and ORC1â5(1âN) to a mix of DNA and ATP in assay buffer.
A 20âµl reaction with full-length proteins typically contained 4ânM (27.5ângâµlâ1) plasmid DNA (10.6âkb), 2âmM ATP, 60ânM MCM2â7, 150ânM ORC6, 120ânM ORC1â5, 150ânM CDC6, and 150ânM CDT1. MCM2â7 was diluted to 10à working concentration in assay buffer. ORC6 was diluted to 20à working concentration in assay buffer + 200âmM sodium chloride. A 20à OCC mix of ORC1â5, CDC6 and CDT1 was prepared in assay buffer + 650âmM sodium chloride. Reactions were started by adding MCM2â7, ORC6, and the OCC mix to a mix of DNA and ATP in assay buffer.
In experiments that contained truncated proteins as well as their full-length counterparts, ORC1â5 and ORC1â5(1âN) were both used at 120ânM and the 20Ã OCC mix (650âmM sodium chloride) method was used. When proteins were omitted, a salt-containing buffer was added instead to maintain the same salt concentration throughout the experiment.
Reactions were started in 2-min intervals and incubated at 37â°C in a thermomixer with shaking at 1,250ârpm. After 30âmin of MCM loading, 2âµl of benzonase nuclease (Sigma-Aldrich, E1014) was added and the mix incubated at 37â°C with shaking for either 0.5âmin or 20âmin depending on the experiment. Then, 20âµl of the mix was transferred to a tube containing 10âµl 3à Stop buffer (assay buffer + 100âmM EDTA, 500âµM proteinase K (Sigma-Aldrich, 107393), 1% SDS) and the mix incubated at 37â°C for 20âmin with shaking.
The sample was diluted with TE (10âmM Tris/HCl pH 8.0, 1âmM EDTA) to 200âµl and an equal volume of phenol/chloroform/isoamyl alcohol (25/24/1; Invitrogen UltraPure, 15593031) was added. The sample was vortexed for 1âmin, transferred to a 5PRIME Phase Lock Gel Heavy spin column (VWR, 733-2478), and spun for 5âmin at 20,000g. The aqueous phase was transferred to a new tube, 20âµl of 3âM sodium acetate pH 5.2, 1âµl of 20âmgâmlâ1 glycogen (Thermo Scientific, R0561), and 550âµl ethanol were added and DNA precipitated overnight at â20â°C. The DNA was pelleted (20,000g, 4â°C, 40âmin), washed with 80% ethanol (20,000g, 4â°C, 10âmin), and air-dried. The pellet was resuspended in 3âµl TE. Then, 1.5âµl 20% Ficoll 400 (Sigma-Aldrich, F2637) was added, and samples were loaded on a pre-run 4â20% Novex TBE gel (Invitrogen, EC62252BOX). The gel was run using TBE running buffer (Invitrogen, LC6675) at 150âV for 50âmin, stained with SYBR Gold (Invitrogen, S11494) for 30âmin, and imaged using an Amersham ImageQuant 800 imager.
Salt stability experiments
For the salt stability experiment in Fig. 1j, the nuclease footprinting assay was performed with the following modifications. MCM loading reactions (20âµl) were set up in assay buffer and were incubated (37â°C, 30âmin, 1,250ârpm). Then, 10âµl of 3à sodium chloride in 1à assay buffer solutions were added to achieve the indicated sodium chloride concentrations, and the reaction was incubated (37â°C, 15âmin, 1,250ârpm). The reactions were diluted to a volume of 200âµl with solutions that adjusted all reaction buffers to 25âmM HEPES/KOH pH 7.6, 20âmM sodium acetate, 150âmM sodium chloride, 10âmM magnesium acetate, 1âmM DTT. Immediately, 10âµl of benzonase nuclease (Sigma-Aldrich, E1014) was added and the mix incubated (37â°C, 0.5âmin, 1,250ârpm). Then, 200âµl were transferred into 100âµl 3à Stop buffer and the mix incubated (37â°C, 20âmin, 1,250ârpm). The samples were extracted with an equal volume of phenolâchloroformâisoamyl alcohol. The aqueous phase was diluted 3-fold with TE, and DNA precipitated with 50% isopropanol, 200âmM sodium chloride, 20âµg glycogen, followed by an 80% ethanol wash. The salt stability experiments in Extended Data Fig. 1e,f were performed in a similar way with the following modifications. MCM was loaded for 30âmin in a single large reaction and split into two tubes before adding assay buffer or a 3à sodium chloride in 1à assay buffer solution to achieve the indicated salt concentration. Samples were taken at the indicated timepoints, diluted, treated with benzonase, and transferred into Stop buffer. For the salt stability experiments in Fig. 3g and Extended Data Fig. 1g, MCM was loaded using the indicated reaction conditions for 30âmin. Then, assay buffer or NaCl-containing assay buffer was added to achieve the indicated salt concentration. After 15âmin incubation, samples were diluted, treated with benzonase for 0.5âmin and transferred into Stop buffer.
Geminin inhibition experiments
For the geminin inhibition experiment shown in Fig. 1i, geminin and CDT1 were mixed on ice prior to setting up the MCM loading reactions. Geminin/CDT1 mixes were prepared at 20à concentration in assay buffer + 200âmM sodium chloride. Reactions were then started by adding MCM (10Ã, assay buffer), ORC6 (20Ã, 200âmM NaCl), geminin-CDT1 mix (20Ã, 200âmM NaCl), and ORC1â5/CDC6 mix (40Ã, 650âmM NaCl) to a mix of DNA and ATP in assay buffer. For the ATPγS timecourse experiment in Extended Data Fig. 1m geminin was added at minute 9 at 300ânM (twofold excess over 150ânM CDT1(âN)).
Nuclease footprinting assay with yeast proteins
Nuclease footprinting experiments with yeast proteins were performed at 30â°C instead of 37â°C. Reactions with yeast proteins contained 4ânM (27.5ângâµlâ1) pJY22 plasmid DNA (10.6âkb, caesium chloride purified), 2âmM ATP, 100ânM Mcm2â7/Cdt1, 40ânM yCdc6 and 40ânM Orc1â6. In experiments containing human and yeast reactions, both were carried out at 30â°C.
Experiments with human origin sequences
A 2,398âbp fragment of the origin at the human lamin B2 (LMNB2) locus, and a 2,398âbp fragment of the origin at the human MYC locus were cloned into the vector pBIG1c. The LMNB2 origin fragment was amplified from human genomic DNA by PCR using oligonucleotides hOri-LamB2-2.4_for (AACGCTCTATGGTCTAAAGATTTACTCAGCAGCCCGGTG) and hOri-LamB2-2.4_rev (AACCCCGATTGAGATATAGATTTTGAGAATTGAGTCTTTGGAAACACTAAG). The MYC origin fragment was amplified using oligonucleotides hOri-Myc_for (AACGCTCTATGGTCTAAAGATTTAAGCTTGTTTGGCCGTTTTAGGG) and hOri-Myc_rev (AACCCCGATTGAGATATAGATTTCTCGAGGCAGGAGGGGAG). The fragments were inserted into pBIG1c that was linearized with the restriction enzyme SwaI using Gibson assembly, and the constructs were sequence verified. Doubly biotinylated DNA fragments of 2,398âbp size were generated by PCR using 5â²-biotinylated oligonucleotides (Integrated DNA Technologies): LamB2 ori using Bio-LamB2-2.4_for ([5â²-biotin]ACTCAGCAGCCCGGTG) and Bio-LamB2-2.4_rev ([5â²-biotin]TGAGAATTGAGTCTTTGGAAACACTAAG), Myc ori using Bio-Myc-2.4_for ([5â²-biotin]AAGCTTGTTTGGCCGTTTTAGGG) and Bio-Myc-2.4_rev ([5â²-biotin]CTCGAGGCAGGAGGGGAG), yeast Ars1 using Bio-Ars1-2.4_for ([5â²-biotin]GGTGGAGATATTCCTTATGGCATG) and Bio-Ars1-2.4_rev ([5â²-biotin]GTAATTCGACCATTCCGACACAG) on pJY22, no origin (pET21a backbone) using Bio-pET21a-2.4_for ([5â²-biotin]CCACAGGTGCGGTTGC) and Bio-pET21a-2.4_rev ([5â²-biotin]TTCACCGTCATCACCGAAAC) on pET21a. The PCR products were column purified (QIAquick PCR purification kit, Qiagen) and purity confirmed by agarose gel electrophoresis. For the experiments shown in Extended Data Fig. 1h,i nuclease footprinting assays were performed using 4ânM doubly biotinylated PCR products and 400ânM streptavidin (Thermo Fisher, 434301).
DNA templates for electron microscopy experiments
The DNA substrate used for electron microscopy imaging of wild-type MCM loading was modified from the pGC209Â (ref. 44) construct, containing two inverted ACS sequences spaced by 70âbp, by adding Widom601 and Widom603 strong positioning sequences at both ends. The Widom sequences map 7 and 5 base pairs away from the inverted ACS sites, making the nucleosome-free region 148 base pairs long. For the experiment shown in Extended Data Fig. 1a, the pGC211Â (ref. 44) construct, which has two inverted ACS sequences spaced by 90âbp, was modified to include Widom601 and Widom603 strong positioning sequences on both ends. The nucleosome-free region is 168 base pairs long in this construct. For MCM5(AG) mutant, Widom sequences were swapped for a suicide substrate for covalent M.HpaII methyltransferase binding (equally efficient at blocking double hexamer sliding28). The plasmids were synthesized by Eurofins and used for PCR amplification with the primer pairs NCP F/NCP R (for nucleosome reconstitution) or Gid70-MTRB F/Gid70-MTRB R (for methyltransferase capping).
Reconstitution of nucleosomes with yeast histones and preparation of HpaII-flanked origins was carried out as described28. In brief, amplified templates were purified by anion exchange chromatography on a 1âml RESOURCE Q column (Cytiva), followed by ethanol precipitation. DNA pellets were resuspended in TE buffer, mixed with purified yeast histone octamers and subjected to dialysis with decreasing NaCl concentration to reconstitute nucleosomes28,45. The chromatinized construct was purified by size exclusion chromatography using a Superose 6 Increase 3.2/300 column (Cytiva). For the methyltransferase construct, DNA was incubated with M.HpaII in 1:6 molar ratio at 30â°C overnight in buffer 1 (50âmM potassium acetate, 25âmM Tris pH 7.5, 10âmM magnesium acetate, 1âmgâmlâ1 bovine serum albumin (BSA), 150âμM S-adenosyl-methionine (NEB)). The M.HpaIIâ70bpâM.HpaII construct was isolated by anion exchange chromatography using a 1âml RESOURCE Q column (Cytiva).
NCP F, 5â²-(Des)-CGATAGAACTCGGGCCGCCCTGGAGAATCGCGGTGCCG-3â²; NCP R, 5â²-CCTGCACCCCAGGGACTTGAAGTAATAAGGAC-3â²; Gid70-MTRB F, atatatCC*GGcctgtATCTCGATTTTTTTATGTTTAGTTTCGC; Gid70-MTRB R -TGGGCGCC*GGAACTGGGTGCTGTaTTTTTATGTTTAGTTCG; (Des), Desthiobiotin TEG; C*, 5-fluoro-2â²-deoxycytosine.
Human MCM loading for cryo-EM
For the hDH loading reaction, 45ânM of chromatinized DNA (nucleosomeâGid70ânucleosome) were incubated with 120ânM ORC1â5(1âN), 120ânM ORC6, 150ânM CDC6(âN), 150ânM CDT1(âN) and 60ânM MCM2â7 in EM buffer (25âmM HEPES-KOH pH 7.6, 100âmM potassium glutamate,10âmM magnesium acetate, 1âmM DTT, 2âmM ATP) resulting in a final volume of 35âµl. Incubation was carried out for 30âmin at 37â°C and 1,250ârpm constant mixing.
The MCM recruitment reaction was established by substituting ATP with ATPγS and a chromatinized origin concentration of 70ânM.
Negative-stain electron microscopy reactions and imaging
The MCM loading reaction for the negative-stain experiment shown in Extended Data Fig. 1a was performed similarly as described above, but using 20ânM ORC1â5(1âN), 20ânM ORC6, 20ânM CDC6(âN), 40ânM MCM2â7, 40ânM CDT1(âN), 7.5ânM of chromatinized DNA (nucleosomeâGid90ânucleosome) in a total volume of 20âµl. Dropout experiments shown in Supplementary Fig. 1 were carried out with the same sample concentrations as described for cryo-EM, but using a final volume of 20âµl per reaction, and omitting one factor at a time. Reactions were diluted twofold (Extended Data Fig. 1a) and 4-fold (Supplementary Fig. 1) in EM buffer where nucleotide was omitted. 300-mesh copper grids coated with a layer of continuous carbon (EM Resolutions, C300Cu100) were glow-discharged at 25âmA for 1âmin using a GloQube Plus Glow Discharge System (Quorum), before applying 4âµl of the sample for 2âmin. Grids were stained with two successive applications of 4âµl of 2% (w/v) uranyl acetate solution. Excess stain was blotted after 40âs using filter paper. Micrographs were collected using a FEI Tecnai G2 Spirit transmission electron microscope operated at 120âkeV, equipped with a 2âK x 2âK GATAN UltraScan 1000 CCD camera. Data collection was carried out at a nominal magnification of 30,000Ã, yielding a pixel size of 3.45âà at the specimen level, and a defocus range of â0.6 to â1.4âµm.
Further electron microscopy investigation of MCM loading with and without ORC6 was performed as follows. Reactions were set up by mixing 45ânM of a M.HpaII-Gid70-M.HpaII DNA template with 90ânM ORC1â5(1âN), 150ânM CDC6(âN), 150ânM CDT1(âN), 60ânM MCM2â7 in EM buffer containing either 2âmM ATP or 2âmM ATPγS, with or without 150ânM ORC6. Samples were incubated at 37â°C for 30âmin under agitation, diluted 1:4 in EM buffer and immediately used for negative staining as described above. Grids were imaged on a FEI Tecnai G2 Spirit microscope using a RIO16 camera at a pixel size of 3.1âà per pixel. Particles were picked using crYOLO46 and extracted with a box size of 144 pixels in Relion 447. After 2 rounds of 2D classification, well-averaging particles were counted, class populations were visualized in a 10âÃâ10 dot plot in Prism10.
Negative-stain image processing
Negative-stain images were processed using Relion 3.147. Particles were picked using Topaz v0.2.548. Contrast transfer function (CTF) parameters were estimated using Gctf v1.0649. Extracted particles were then subjected to reference-free 2D classification.
Cryo-EM reactions and imaging with ORC6
UltrAuFoil R1.2/1.3 300-mesh grids (Quantifoil) were glow-discharged at 40âmA for 5âmin using a GloQube Plus Glow Discharge System (Quorum), before applying 3âµl graphene oxide dispersion (10âml graphene oxide flake dispersion (Sigma) diluted in 80âml water; aggregates removed by centrifugation at 500g for 1âmin). Incubation was carried out for three minutes, followed by blotting of excess liquid and three successive washes with 20âµl droplets of water. After 1â2âh drying at room temperature, 4âµl of the undiluted (ATP) reaction or the 3:1 diluted (ATPγS) reaction were applied to grids for 60âs at room temperature and 90% humidity in a Vitrobot Mark IV (Thermo Fisher). Grids were double-side plotted with force 0 for 5âs and immediately plunge frozen in liquid ethane. Micrographs were collected in counting mode using a pixel size of 1.08âà on a Titan Krios transmission electron microscope with a K2 Summit direct electron detector and BioQuantum energy filter. A total electron dose of 49.28âeââà â2 was used over 32 dose-fractioned movie frames and a total exposure time of 9.4âs. The defocus ranged from â1.0 to â2.5âµm. 3,589 movies were collected for the ATP reaction and a total of 31,569 movies for the ATPγS reaction.
Cryo-EM reactions and imaging without ORC6
Gid70 DNA template (45ânM), carrying a TwinStrep-tagged M.HpaII roadblock at each end, were mixed with 90ânM ORC1â5(1âN), 150ânM CDC6(âN), 150ânM CDT1(âN) and 60ânM MCM2â7 in EM buffer to a total volume of 40âµl, and incubated for 30âmin under agitation at 37â°C. Four microlitres of 50% diluted loading reaction were applied onto UltrAuFoil R1.2/1.3 300-mesh grids, which were coated with a graphene oxide layer as described above. After one minute of on-grid incubation in a Vitrobot Mark IV, the grids were blotted from both sides with Whatman Filter paper (blotting strength 0, blot time 3.5âs) and plunged into liquified ethane. Grids were clipped and stored in liquid nitrogen prior to data collection.
hDH loading reactions without CDC6 imaged
A hDH loading reaction was prepared by co-incubating 45ânM of a M.HpaII-Gid70-M.HpaII DNA with 90ânM ORC1â5(1âN), 150ânM CDT1(âN) and 60ânM MCM2â7 in assay buffer containing 60âmM sodium acetate (supplemented with 2âmM ATP) for 30âmin at 37â°C under agitation. The sample was diluted 1:4 and subsequently negative-stained as described above. One hundred and fourteen micrographs were acquired using a RIO16 detector on a FEI TECNAI G2 Spirit Microscope at 3.1âà per pixel. 69,785 particles were extracted with a 144-pixel box after crYOLO46 picking and submitted to multiple rounds of 2D classification to identify MCM hDH particles in Relion 447.
Cryo-image processing of hDH loading
Image processing was performed using Relion 4.0b-GPU and cryoSPARC v3.3.250 at different stages of the processing pipeline as indicated in Extended Data Fig. 2. Beam-induced motion was accounted for by the Relion implementation with 5âÃâ5 patches and CTF parameters were estimated using CTFFIND v4.1.1051. Particle picking was carried out using Topaz v0.2.448, followed by particle extraction with a 440-pixel box and rescaling to 110 pixels in cryoSPARC. Three thousand, five hundred and eighty-nine micrographs with 970,326 particles were selected based on the CTF fit resolution of 2.57â4.50âà , CTF fit cross-correlation of 0.07â0.27 and median pick score of 20.18â43.55. Three rounds of reference-free 2D classification, ab initio reconstruction and heterogeneous refinement were conducted to identify 49,485 hDH particles. After particle extraction without downscaling and using a 400-pixel box, the particle stack was further cleaned up by 2D classification, which yielded 19,049 particles used to compute high-resolution hDH 2D class averages. Ab initio reconstruction, followed by homogeneous, non-uniform and local refinement with C2 symmetry resulted in a map with 3.1âà resolution. The particle stack was re-extracted, re-grouped, and cleaned from duplicate particles using Relion. Fifteen thousand, eight hundred and seventy-four particles of the highest quality were isolated by 3D classification without alignment using a 320âà mask and a regularization parameter T of 4. Particles were polished47 subjected to 2D classification without alignment and 3D refinement imposing C2 symmetry. CTF parameters were optimized47 in three rounds (first, per-particle defocus, per-micrograph astigmatism; second, per-particle defocus, per-particle astigmatism, beamtilt; third, per-particle defocus, per-particle astigmatism, beamtilt, trefoil, 4th order aberrations) to yield a 3.3âà resolution hDH structure. Homogeneous and non-uniform refinement with C2 symmetry in cryoSPARC resulted in the final map at 3.1âà resolution.
To determine the structure of hSH from the same dataset, a new Topaz model was trained. One hundred and seventy-eight thousand, eight hundred and fifty three particles were extracted with a 440-pixel box, downscaled to 110 pixels. Smaller particles and contaminations were removed by 2D classification in cryoSPARC. Initial volumes were obtained by ab initio reconstruction with six classes. Thirty-seven thousand, three hundred and ninety-six hSH particles were then isolated in two rounds of heterogeneous refinement (Extended Data Fig. 7a) and re-extracted with a 400-pixel box. Homogeneous, non-uniform and local refinement with a mask encompassing the entire hSH yielded a 3.4âà resolution structure. The same particles were re-extracted and 3D refined in Relion. CTF refinement (per-particle defocus, per-micrograph astigmatism), Bayesian polishing and one additional round of CTF refinement (per-particle defocus, per-particle astigmatism, beamtilt) were carried out.  Twenty-five thousand and sixty-nine high-resolution hSH particles were isolated by 3D classification. Homogeneous, non-uniform and local refinement with a mask around the hSH yielded 3.2âà resolution in cryoSPARC.
Cryo-image processing of MCM recruitment
Movies were corrected for beam-induced motion using the Relion implementation with 5âÃâ5 patches in Relion 4.0b-GPU52 and CTF parameters were estimated using CTFFIND v4.1.1351. A Topaz model48 was trained and 1,334,277 particles were picked from 31,569 micrographs. Particles were extracted with a 416-pixel box, rescaled to 104 pixels. Six hundred and twenty-nine thousand, two hundred and forty-one MCM-containing particles (that is, hOCCM, hSH and hMO*) were isolated using reference-free 2D classification. Initial volumes were generated using ab initio reconstruction in cryoSPARC. Low-pass filtered volumes were used for multi-reference 3D classification in Relion (Extended Data Fig. 5). Particles contributing to the different complexes (114,995 hSH, 170,792 hOCCM and 203,088 hMO* particles) were re-extracted using a 400-pixel box without rescaling. Homogeneous, non-uniform and local refinements with masks around the entire respective complex resulted in 3.6âà resolution structure of the hSH, 4.0âà hOCCM and 3.7âà hMO* in cryoSPARC. Bayesian polishing was carried out for each particle stack individually in Relion47.
The hOCCM was then subjected to three rounds of CTF refinement (first, per-particle defocus, per-micrograph astigmatism; second, anisotropic magnification; third, per-particle defocus, per-particle astigmatism, beamtilt) followed by another round of Bayesian polishing in Relion. A mask around CDC6 and portions of ORC1â5 was generated to carry out focused 3D classification without alignment. This yielded 100,567 particles with good density for ORCâCDC6. 3D refinement in Relion was followed by homogeneous refinement in cryoSPARC. Local refinement with a mask around ORCâCDC6 was used to improve alignment on this part of the complex. Thirty-four thousand, one hundred and sixteen hOCCM particles with well-resolved CDC6 density and 49,771 hOC1M particles that lacked CDC6 were isolated by 3D classification without alignment in cryoSPARC. Both hOCCM and hOC1M structures were locally refined to 3.8 and 4.1âà , respectively, using a mask encompassing the entire complex.
Complexes containing single-loaded hexamers, hSH and hMO* particles were initially processed together (Extended Data Fig. 5). Three-dimensional refinement in Relion using a mask around the MCM yielded a 3.8âà hSH structure. Two rounds of CTF refinement (first, per-particle defocus, per-micrograph astigmatism; second, anisotropic magnification) and another round of Bayesian polishing was carried out. One hundred and thirty-five thousand, seven hundred and forty-two hSH particles and 182,341 hMO* particles were subsequently separated by multi-reference 3D classification using the initial maps of hSH and hMO*, low-pass filtered to 30âà . The hSH was refined in cryoSPARC (homogeneous, non-uniform, followed by local refinement) to 3.4âà . Homogeneous and non-uniform refinement of the hMO* resulted in a consensus map solved to 3.6âà resolution. The MCM portion of the complex was locally refined to 3.5âà , while the ORC was refined to 4.0âà .
Cryo-EM of hDH loading without ORC6
A total of 5,158 movies were collected in counting mode on a 200âkV Talos transmission electron microscope using a pixel size of 1.61âà per pixel with a total dose of 48âeââà â2 and a defocus range of â2 to â3.5âμM (step size 0.25âμM). Movies were corrected for beam-induced motion with the Relion implementation with 5âÃâ5 patches in Relion 5.0 and CTF parameters estimated with CTFFIND v4.1.1351. Subsequent micrograph curation reduced the number of micrographs to 2,168. Four hundred and fifty-six thousand, six hundred and eighty-six particles were picked using template matching in cryoSPARC v4.4.150 and extracted at a pixel size of 3.22âà per pixel (2à binned) and subjected to two rounds of 2D classification. A total of 77,002 particles were used to generate an ab initio reconstruction in C1. Particles were re-extracted at full resolution, refined and then symmetry expanded in C2. Three-dimensional classification was performed using 4 classes. The class with the highest quality DNA density containing 74,880 particles was subject to a local refinement resulting in a 4.1âà structure according to gold-standard FSC at 0.143 criterion.
MCM5(AG) DH loading for cryo-EM
A 50-µl MCM DH loading reaction was assembled using the protocol described in âHuman MCM loading for cryo-EMâ. In short, 45ânM M.HpaIIâGid70âM.HpaII DNA capped at each end with TwinStrep-tagged M.HpaII, was co-incubated with 90ânM ORC1â5(1âN), 150ânM CDC6(âN), 150ânM CDT1(âN) and 60ânM MCM5(AG)âMCM2â7 in EM buffer at 37â°C under agitation. After 30âmin, the loading reaction was diluted either by 40% or 75% with EM buffer and immediately used for cryo-EM.
UltrAuFoil R1.2/1.3 300-mesh grids were coated with a graphene oxide support as described above. Four microlitres of diluted loading reaction were applied onto each grid in a Vitrobot Mark IV set to 22â°C and 90% humidity. After one minute on-grid incubation, grids were double-side blotted for 4.5âs with blot force 0 and plunged into liquid ethane. Grids were subsequently clipped and stored in liquid nitrogen until data collection.
Cryo-EM of MCM5(AG) DH
In total, 13,203 movies were acquired from two grids at 92,000à magnification (1.58âà per pixel) on a Glacios microscope equipped with a Falcon 3 direct electron detector operated in linear mode. A total dose of 50âeââà â2 (exposure time 1.12âs) and a defocus range of â1 to â2.5âµm (0.3âµm step size) were applied. Movie frames were aligned using Relion 4.0 (ref. 52) and CTF was estimated using GCTF v1.06 (ref. 49). Two million, eight hundred and seventy-five thousand, eight hundred and twenty-five particles were picked using a pre-trained Topaz network48, extracted with a box size of 64 pixels (4à binned to 6.32âà per pixel) and imported in cryoSPARC v4.050 for 2 rounds of 2D classification. After 2D cleaning, two ab initio models were generated with C1 symmetry using a subset of 50,000 particles. All 379,664 particles were subjected to one round of heterogenous refinement against the two ab initio reconstructions, followed by homogenous refinement in C1. Particles were re-extracted in Relion with a box size of 300 pixels (unbinned at 1.58âà per pixel) and 2D-classified in cryoSPARC. The resulting 322,548 particles were used to generate an unbinned ab initio reconstruction in C1, yielding a 5.76âà map after non-uniform refinement in C1. Three-dimensional classification in Relion without alignment using a regularization parameter T of 200 distributed the particles into four classes with roughly equal populations, differing by presence of both, either or no MCM6 WH domain. The best-resolved class (showing density for both MCM6 WH domains) was non-uniform and locally refined in cryoSPARC with C2 symmetry, yielding a final resolution of 5.6âà according to gold-standard FSC and the 0.143 criterion. Local resolution estimation was carried out in cryoSPARC. Refinement statistics are reported in Extended Data Table 1.
hDH model building
AlphaFold-Multimer53 was used to generate models of the ATPase tier (including the WH domains) of the hexameric human MCM2â7 assembly as well as the amino-terminal tier. Each model was rigid-body docked into one hexamer of the 3.1-Ã resolution hDH map using UCSF ChimeraX v1.6.1Â (ref. 54). Each chain was refined in Coot v0.9.8.1 EL55 and sections that could not be confidently built were deleted54. The models of the ATPase and amino-terminal tiers were then combined. ATP, ADP, magnesium and zinc ions were added in the pertinent sites. Idealized B-form DNA was first docked into the density of the double helix and then manually modified to account for the stretch of underwound and melted DNA within the N-terminal dimerization interface. The model consisting of the MCM2â7 hexamer, ligands and DNA was adjusted using ISOLDE 1.6.0Â (ref. 56), imposing ligand, secondary and base pairing restraints. This was followed by an iterative process of real space refinement with restraints on geometry, secondary structure, metal coordination and nucleic acid planarity in Phenix v1.21 (ref. 57) and manual adjustments in Coot and ISOLDE. To generate the hDH, a copy of the refined MCM hexamer was generated and rigid-body docked into the second hexamer of the hDH map. Clashes at the interface of the two hexamers were addressed using ISOLDE. The resulting model was subjected to real space refinement in Phenix. Refinement statistics are reported in Extended Data Table 1.
hSH model building
The atomic model of the hDH was rigid-body docked into the 3.2âà cryo-EM map of the hSH using UCSF ChimeraX v1.6.1 (ref. 58). The second hexamer was deleted, nucleotides were inspected and the DNA was replaced by an idealized B-form duplex DNA using Coot v0.9.8.1 EL59. The model was refined with ISOLDE 1.6.0 (ref. 56) and Phenix real space refinement with restraints on geometry, secondary structure, metal coordination and nucleic acid planarity57. Refinement statistics are reported in Extended Data Table 1.
hMO* model building
To generate the model of the hMO* complex, the atomic structure of the hSH described above was docked into the map of the globally refined hMO* map using UCSF ChimeraX v1.6.1 (ref. 54). The atomic model of H. sapiens ORC6 was retrieved from the AlphaFold Protein Structure Database (accession code AF-Q9Y5N6-F1). To guide the positioning of ORC6, the yMO (PDB entry 6RQC)28 was aligned with the human MCM in the map. Based on the alignment with the S. cerevisiae Orc6, the N-terminal cyclin box domain (residues 1â94) of human ORC6 was positioned next to the N-terminal domains of MCM2 and MCM6. The domain was then rigid-body docked into the density. The second cyclin box domain (residues 95â190) was docked into the adjacent density of the map that is positioned between MCM6 and ORC5. Docking solutions with the highest cross-correlation scores were chosen. The starting model of ORC1â5 was the open conformation of the human ORC1â5 complex (PDB code 7JPR)60. The model was docked into the locally refined map of the ORC at 4.0âà resolution. The DNA model of the yMO complex (PDB entry 6RQC)28 was fit into the DNA density in the globally refined hMO* map. The DNA model was refined in Coot v0.9.8.1 EL59 applying all-molecule self-restraints 6.0 and DNA B-form restraints, before combining it with the DNA in the hSH. ORC6 was combined with the MCM and DNA into one PDB model. The two models of ORC6âMCM and ORC1â5 were iteratively refined with ISOLDE 1.6.0 (ref. 56) and Phenix real space refinement with restraints on geometry, secondary structure, metal coordination and nucleic acid planarity57 against the globally refined map of hMO* and the locally refined map of the ORC, respectively. For illustration purposes, a composite map was generated using Phenix and the refined ORC6âMCM and ORC1â5 models were combined into one PDB model. Refinement statistics are reported in Extended Data Table 2.
hOCCM model building
To assemble the atomic model of hOCCM, the human ORC1â5âDNA model (PDB entry 7JPS), the ORC2 WH domain of the ORC1â5 model (PDB entry 7JPR)60 and the prediction of human CDC6 retrieved from AlphaFold Protein Structure Database (accession code AF-Q99741-F1) were rigid-body docked into the 3.8âà cryo-EM map using UCSF ChimeraX v1.6.1 (ref. 54). Unstructured parts of the models, which were not visible in the hOCCM map, were deleted. The hSH model was docked into the map and each subunit was split and fitted independently into the density. The N-terminal tiers of MCM7, MCM3 and MCM5 were deleted from the atomic coordinates file as the local quality of region of the map was deemed insufficient for model building. The positioning of the ATPase tiers of the same MCM subunits was guided by the position in the hSH and no further adjustment of atomic positions was carried out due to the limited local resolution. The atomic models of the MCM3 and MCM7 WH domains were overlayed with equivalent domains in yOCCM. The atomic model of CDT1 was retrieved from the AlphaFold database (accession code AF-Q9H211-F1) and the three structured domains (residues 167â387, 418â440 and 441â546) were extracted from the model. Each domain was overlayed with yeast Cdt1 in yOCCM (PDB entry 5V8F)29. All models were combined into one PDB model and adjusted using Coot v0.9.8.1 EL59 and ISOLDE 1.6.0 (ref. 56). Models of ATPγS and magnesium were added in the ATPase active sites of MCM2â6, MCM6â4, ORC1â4, ORC4â5 and ORC5â3. A 39-mer idealized B-form duplex DNA was generated and fit into the density by applying all-molecule self-restraints 6.0 and DNA B-form restraints in Coot. The DNA coordinates were then merged with the hOCCM model before carrying out Phenix real space refinement with restraints on geometry, secondary structure, metal coordination and nucleic acid planarity57. Refinement statistics are reported in Extended Data Table 3.
Analysis of proteinâDNA contacts
ProteinâDNA contacts were analysed using the DNAproDB web-based visualization tool61,62.
Statistics and reproducibility
Proteins were independently purified at least twice (Fig. 1a). The experiments in Figs. 1d,f,g,j, 2e,g and 3a,f, Extended Data Fig. 1a,n and Supplementary Fig. 1aâd were performed three times. The experiments in Figs. 1c,e,h,i and 3g,i,k, Extended Data Figs. 1bâi,m, 3d and 4a and Supplementary Figs. 1eâh and 2b were performed twice. The micrographs shown in Extended Data Figs. 2a and 4b are representative micrographs of cryo-EM datasets. Similar experiments have been analysed multiple times by negative-stain electron microscopy.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.