Mice
All of the mice used in this study were on the B6 genetic background. WT C57BL/6J mice were purchased from TACONIC. Nr1h3fl/flNr1h2fl/fl mice, provided by J.-à . Gustafsson (Karolinska Institutet), were crossed with Villin-cre mice (B6.SJL-Tg(vil-cre)997Gum/J, provided by J.-à . Gustafsson; and B6.Cg-Tg(Vil1-Cre)1000Gum/J, obtained from Jackson laboratory) to generate Nr1h3fl/flNr1h2fl/fl and Villin-cre;Nr1h3fl/flNr1h2fl/fl littermate controls. Cyp27a1â/â (B6.129-Cyp27a1tm1Elt/J) mice were purchased from Jackson Laboratory and bred locally as Cyp27a1+/â à Cyp27a1+/â or Cyp27a1+/â à Cyp27a1â/â breeding pairs. Lgr5-eGFPIRES-creERT2 mice50 were maintained on a C57BL/6J background. Villin-creERT2 Yap1fl/flTazfl/fl and Yap1fl/flTazfl/fl control mice were provided by H. L. Larsen (University of Copenhagen). LXR-sufficient and Cyp27a1-sufficient controls for deficient mice were littermates and co-housed (unless specifically indicated). Mice, both males and females, were generally used between 8 and 20 weeks of age. Mice were maintained under specific-pathogen-free conditions at Karolinska Institutet, except for some experiments with Cyp27a1â/â and littermate controls in which some of the mice were housed in an MPV-positive animal facility (the experiments with DSS administration and half of the irradiation experiments performed on Cyp27a1â/â and littermate controls). ApcMin/+ mice (C57BL/6J-ApcMin/J), provided by S. Huber (some of the ApcMin/+ mice were on a Hmgb1-floxed background without any endogenous Cre), were maintained under specific-pathogen-free conditions at Hamburg-Eppendorf University (Hamburg). Aregâ/â mice were provided by M. Biton and were maintained under specific-pathogen-free conditions at Weizmann Institute of Science (Israel). When administered in vivo, GW3965 (Adooq Bioscience) was administered through formulated drug in diet at 50âmg per kg per day (Research Diet and SSNIFF). The same purified based diet (D11112201, Research Diet or AIG93G + Inulin, SSNIFF), used to formulate GW3965-diet, was used in standard-diet fed mice. BrdU was prepared at 10âmgâmlâ1 in PBS, sterile filtered through a 0.22âµm filter and injected at 100âmg per kg body weight 2âh before euthanasia. No randomization or blinding was used. Sample sizes were determined based on pilot and preliminary experiments. All of the experimental procedures were performed in accordance with national and institutional guidelines and regulations. Animals used for experiments performed at Karolinska Institutet (Stockholm, Sweden) were maintained under specific-pathogen-free conditions at Karolinska Institutet (Stockholm, Sweden) animal facilities and were handled according to protocols approved by the Stockholm Regional Ethics Committee.
Colitis model
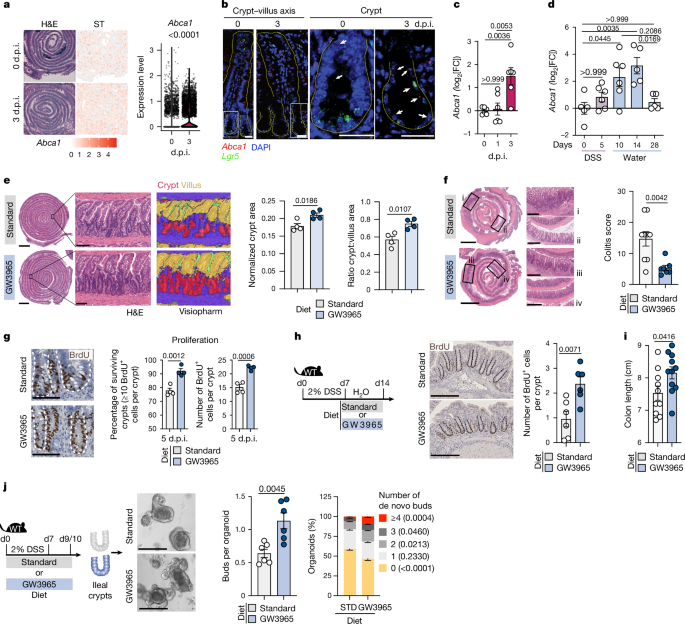
Colitis was induced by administration of 2â3% (w/v) DSS (TdB Consultancy, molecular weight 40 kDa) dissolved in drinking water ad libitum for 6â7âdays, followed by regular drinking water. Body weight loss and mouse health status were monitored regularly. At the indicated timepoints, the colon length was measured, 0.5âcm of distal colonic biopsies was collected for RNA extraction (when indicated) and the colonic tissue was cut open longitudinally, Swiss-rolled and fixed in 10% buffered formalin for histological analysis. For ileal organoids and gene expression analyses in ileal crypts after DSS administration, distal third of the SI (that is, ileum) was collected at the indicated timepoints and processed for crypt isolation as described below in the âSI organoidsâ section. The colitis score was assessed in a blinded manner in paraffin-embedded sections stained with H&E and calculated using a TJL-based system according to the following formula: (degree of severityâ+âdegree of hyperplasiaâ+âdegree of ulceration)âÃâpercentage area involved. Each parameter was scored from 0 to 3 according to the following description: (a) severity: 0, unaffected; 1, single/widely scattered, LP involved; 2, larger or involving submucosa; 3, ulcers longer than 20 crypts; (b) hyperplasia: 0, normal; 1, 1â2à normal thickness; 2, 2â3à normal thickness, hyperchromasy, reduced goblet cells, elongated crypts, increased mitoses; 3, 4à normal thickness, hyperchromasy, reduced goblet cells, elongated crypts, high mitotic index, crypt branching; (c) ulceration: 0, no ulcers; 1, 1â2 ulcers, less than 20 crypts; 2, 1â4 ulcers, 20â40 crypts; 3, >4 ulcers, >40 crypts; (d) percentage of area involved: 0, 0%; 1, 10â30%; 2, 40â70%; 3, >70%.
Mouse tumour models
AOMâDSS model
Mice were intraperitoneally (i.p.) injected with 10âmg per kg of body weight AOM (Sigma Aldrich) resuspended at 10âmgâmlâ1 in water, which was further diluted to 1âmgâmlâ1 in PBS before injecting in mice. Administration of modified diet (standard or GW3965 diet ad libitum) was started on the same day and continued for the entire duration of the experiment. Then, 1âweek later, 1.5â2% (w/v) DSS (TdB Consultancy, molecular weight 40 kDa) was administered in the drinking water for 7âdays followed by 2âweeks of regular water. The same DSS treatment was repeated for two more cycles (three cycles in total), each time with an interval of 2âweeks of regular water in between. Mice were euthanized at the indicated timepoints and colon samples were collected. Anti-CD19 (300âµg per mouse per injection, BioXcell, 1D3) and anti-CD8β (300âµg per mouse per injection, BioXcell, Lyt3.2) were treated alone or in combination i.p. every 7âdays starting from day 22 (that is, after the first round of DSS treatment) until the end of the experiment. Control mice were injected 300âµg per mouse per injection with isotype control (rat IgG2a, BioXcell) or PBS. The tumour number and size were evaluated manually using a digital micro-caliper. Tissue biopsies for RNA extraction were collected from the distal colon (corresponding to tumour areas). Colon tissues were then Swiss-rolled for histological analysis. Histological scoring of tumours was performed in a blinded manner using the H&E stained Swiss-rolls in which the tumour areas were categorized into either well-differentiated, well-moderately differentiated or moderately differentiated. The dysplastic regions seen in the tissues were from the non-tumour area and not the dysplasia within the tumour.
Apc
Min/+ model
ApcMin/+ mice (aged 4â5 weeks) were given the modified diet (standard or GW3965-diet) ad libitum for the entire duration of the experiment. Mice were euthanized at around 4âmonths of age and macroscopic tumours were measured and counted manually.
Total-body irradiation
Adult mice were exposed to a dose of 10âGy from an X Radiation Unit (CIX3 X-Ray Cabinet or with an X-RAD 320 irradiation source with 20âÃâ20âcm irradiation field within a mouse pie) and euthanized at the indicated timepoints. At the indicated timepoints, SI tissues were either Swiss rolled for histological analyses, Visium processing or SI crypts were isolated as described below in the âSI organoidsâ section. Histological analyses of H&E-stained tissues embedded in paraffin was conducted using Visiopharm (v.23.01), an artificial-intelligence-powered pathology program that uses a trainable algorithm based on a convolutional neural network. The regions of interest (ROIs) within the Swiss roll (that is, the total measured area) were manually selected to exclude damaged areas caused by tissue rolling artefacts. Subsequently, the crypt and villus area within these selected ROIs were quantified. Crypt hyperplasia was evaluated using normalized crypt area (that is, crypt area/total area) and the crypt area-to-villus area ratio.
Radiation-induced salivary gland injury
Mice at the indicated timepoints were first anaesthetized in the isoflurane chamber (1% O2, 1% air and 5% flow rate), and, once asleep, were transferred to the irradiator bed with 0.3% O2, 0.3% air and 1.5% flow rate until the irradiation protocol was finished. The mice were irradiated individually using X-ray irradiator CIX3 (Xstrahl) with only the neck exposed using 1âcm focal tube and rest of the body and head shielded. An X-ray irradiation dose of 9âGy (that is, 300âkV, 10âmA for a period of 7.04âmin) was administered. After irradiation, the mice were monitored until they were fully conscious and awake. Non-irradiated control mice were subjected only to the anaesthesia protocol without being irradiated. Mice were euthanized at 14âd.p.i. and salivary glands were collected and fixed in 10% buffered formalin for histological analysis.
Histological quantification of salivary gland tissues
To get automatized quantification of salivary gland histology, a deep learning algorithm was developed to automatically segment and classify ducts, blood and air from histological tissue samples. For dataset preparation, histological images underwent tile extraction and preprocessing, following the NoCodeSeg framework (35155486). The dataset included 43 training patients and 1 testing patient. Tiles of 512âÃâ512 pixels in size were obtained using QuPath. The training dataset comprised 35,506 objects, addressing class imbalances through augmentation. For deep learning model training, a deep learning model, based on the Xception architecture51 (https://arxiv.org/abs/1610.02357), was trained using the MIB52 MATLAB package. Parameters were saved for the model, and training used 512âÃâ512 pixel patches. The validation set was split randomly for training (97%) and validation (3%). For model evaluation, post-training, the modelâs performance was assessed using loss curves and a confusion matrix on the test set. For application of trained model, the trained model was integrated into FastPathology for practical application. This involved copying the model and pipeline files, creating a new project, adding images and selecting the pipeline for processing. Annotations in QuPath were loaded into QuPath by downloading and running the provided script for each image in the project. More details about the training and inference of the model can be found on the GitHub repository (https://github.com/BIIFSweden/EduardoVillablanca2023-1). Outliers were identified and removed from the imported data in R using the boxplot function. Subsequently, the combined GW0 (nâ=â2) and STD0 (nâ=â2) datasets were analysed, yielding summary statistics. Bin analysis was performed using breaks delineated at 0 to first quartile, first quartile to mean, mean to third quartile and third quartile to the maximum, labelled as noise, small, medium and large, respectively. Noise-tagged ducts were filtered out, and their distributions were visualized through violin plots. The mean and s.d. of duct areas were computed for each biological replicate, followed by the application of Welch two sample t-tests to compare treatments.
SI organoids
Mouse SI organoids were obtained from purified crypts derived from the entire SI, unless specified. In brief, SI tissue was flushed with cold PBS, cleaned from attached fat tissue, cut opened longitudinally and subsequently cut into approximately 0.5âcm pieces. Tissues were incubated in 30âml of 10âmM cold PBS-EDTA on ice for the following incubation periods: 10âmin, 3âÃâ15âmin and 1âh. After each incubation, tubes were gently shaken, supernatant containing tissue debris and villi fractions were discarded and fresh PBS-EDTA was added. After the last incubation, the tubes were vigorously shaken and filtered through 70âµm cell strainers and isolated SI crypts were retrieved in the flow-through. The last step was repeated until no more crypts were present in the flow-through. Alternatively, SI tissue, flushed with PBS and cut opened longitudinally, was cut into 4â5 pieces (approximately 7âcm long) and incubated for 1âh in ice cold 10âmM PBS-EDTA. Using two glass slides, intestinal villi were then removed by gentle scraping of the luminal side. SI crypts were then scraped by applying stronger pressure with the glass slides and collected in recipient tubes filled with cold PBS. Crypts were centrifuged at 4â°C, 300g for 5âmin. The basic culture medium (ENR) contained advanced DMEM/F12, 1à penicillinâstreptomycin, 1à GlutaMAX (Thermo Fisher Scientific), 10âmM HEPES (Thermo Fisher Scientific), 1à B27 (Life Technologies), 1à N2 (Life Technologies), 1âmM N-acetylcysteine (Sigma-Aldrich) and was supplemented with 50ângâmlâ1 of murine recombinant epidermal growth factor (that is, EGF, from R&D), 250â500ângâmlâ1 recombinant murine R-spondin (R&D) and 100ângâmlâ1 recombinant murine Noggin (Peprotech). For experiments with NR medium, the same medium as above with the exception of EGF was prepared. SI crypts were resuspended in 30â40% basic culture medium with 60â70% Matrigel (Corning) and 20âµl containing approximately 300â500 crypts were plated in a prewarmed flat-bottom 48-well plate. The plate was placed at 37â°C and allowed to solidify for 15âmin before 200âµl of ENR or NR medium (containing the different stimuli) was overlaid. The medium was replaced every 2âdays with fresh medium and the cultures were maintained at 37â°C in fully humidified chambers containing 5% CO2. Only on the first 2âdays of culture, 10âµM Y-27632 (Sigma-Aldrich) was added in the medium. For in vitro stimulation, LXR agonist GW3965 (1âµM, Sigma-Aldrich) or RGX-104 (1âµM, MedChemExpress) was added in the medium for the entire duration of the organoid culture. DMSO was used as vehicle. For rAREG and anti-AREG experiment, rAREG (50ângâmlâ1, Peprotech) and/or anti-AREG (1.5âµgâmlâ1, R&D Systems) were added in the medium for the entire duration of the organoid culture. For experiments with cholesterol, water soluble cholesterol (50âmM, Sigma-Aldrich) was added into the medium for the entire duration of the organoid culture. For organoids from Villin–creERT2 Yap1fl/flTazfl/fl mice, tamoxifen (1âµM, Sigma-Aldrich) treatment was performed in secondary organoids as indicated with or without GW3965 treatment. In brief, primary organoids from Villin-creERT2 Yap1fl/flTazfl/fl mice grown under either ENR or NR conditions were recovered from Matrigel by two washes with 0.1% BSA PBS. The organoids were mechanically fragmented by passing (30 times) through a 200âμl pipette tip. Equal amounts of organoid fragments were plated in 20âµl Matrigel (70% Matrigel) dome in a prewarmed 48-well plate and cultured for 5â8 days as indicated. Secondary organoids were cultured using either ENR or NR medium containing DMSO or GW3965 (1âµM). Y-27632 (10âµM, Sigma-Aldrich) was added only during the first 2âdays of the culture, with medium change done every 2â3 days afterwards. Tamoxifen (1âμM; Sigma-Aldrich) was added at the indicated timepoints. For ileal organoids after DSS treatment, mice were given 2% DSS in the drinking water for 7âdays followed by normal drinking water. Two to three days after DSS removal, the crypts were isolated from the distal third of the SI (that is, ileum) and organoids were cultured in either ENR or NR medium as described above. Crypt domain budding was quantified under the microscope at the indicated days in a blinded manner. Each condition was plated in triplicates and 2â3 wells per condition were quantified. Each dot in the quantification plot represents one mouse and an average of 2â3 technical replicates. Furthermore, organoids were stratified based on the number of buds/organoid (0, 1, 2, 3 or â¥4) and plotted as the percentage of organoids per well. Organoids were collected at the indicated timepoints and stored in RLT-Plus buffer (Qiagen)â+â2% β-mercaptoethanol (Gibco) at â80â°C until RNA extraction.
Single-cell replating experiment for SI organoids
For replating experiments (passage 1), single cells were isolated from WT (C57BL/6) mouse primary SI organoids grown under either ENR-DMSO, NR-DMSO or NR-GW3965 (1âµM) for a period of 5 days, as described before. In brief, primary organoids from respective culture conditions were digested with TrypLE Express (Gibco) with 1,200âUâmlâ1 of DNase I (Roche) at 37â°C for 10âmin. Cells were then washed; dead cells were removed using the EasySep dead cell removal (Annexin V) kit (STEM Cells), filtered using 40âµm filters and live cells were counted using the Guava Muse cell counter (Cytek) and the Muse Count and Viability Kit (Cytek). Equal numbers (10,000) of live IECs per condition in a 20âµl Matrigel (70% Matrigel) dome were plated in prewarmed 48-well plate and cultured for 6â7 days and were longitudinally imaged using the Incucyte S3 Live-Cell Analysis Instrument (Sartorius). Cells from ENR-DMSO, NR-DMSO or NR-GW3965 (1âµM) were cultured in respective medium with WNT-surrogate Fc (1ânM) (IpA Therapeutics) and Y-27632 (10âµM, Sigma-Aldrich) added only during the first 3 days of the culture, with medium change done every 2 days afterwards. Organoid numbers on day 4 counted using the Incucyte Organoid Analysis Software Module were used to estimate the organoid plating efficiency (that is, the number of organoids/10,000âÃâ100). Buds per organoid and the percentage of budding organoids were estimated from day 6/7 images which were counted manually using Fiji/ImageJ (NIH).
Colon organoids and single-cell replating experiment
After crypt isolation from the whole colon of WT (C57BL/6) mice, crypts were resuspended in 70% Matrigel (BD Bioscience) and 30% basal medium (advanced DMEM/F12, 1à penicillinâstreptomycin, 1à GlutaMAX (Thermo Fisher Scientific), 10âmM HEPES (Thermo Fisher Scientific), 1à B27 (Life Technologies), 1âmM N-acetylcysteine (Sigma-Aldrich)). To establish primary colon organoids, approximately 500â600 crypts embedded in 20âμl of Matrigel (70%):crypt suspension (30%) and were seeded onto each well of a 48-well plate. Once solidified, the Matrigel was incubated in 200âμl WENR stem cell medium for 3âdays supplemented with 10âμM Y-27632 (R&D Systems). The stem cell culture medium (WENR) contained basal medium supplemented with 50ângâmlâ1 of recombinant murine EGF (R&D), 500ângâmlâ1 recombinant murine R-spondin (R&D), 100ângâmlâ1 recombinant murine noggin (Peprotech), 500ânM A83-01 (Sigma-Aldrich), 10âmM nicotinamide (Sigma-Aldrich) and 1ânM WNT surrogate Fc (IpA Therapeutics). On day 3, WENR stem cell medium was replaced with ENR differentiating medium lacking WNT surrogate Fc, nicotinamide and Y-27632, with subsequent medium change every 2âdays. All throughout the primary organoid culture, DMSO or GW3965 (1âµM, Sigma-Aldrich) were added to the culture medium.
For replating experiments (passage 1), single cells were isolated from the primary colon organoids grown under either DMSO or GW3965 (1âµM) for a period of 6â7 days. In brief, primary colon organoids from the respective culture conditions were digested with TrypLE Express (Gibco) with 1,200âUâmlâ1 of DNase I (Roche) at 37â°C for 10âmin. Cells were then washed; dead cells were removed using the EasySep dead cell removal (annexin V) kit (StemCell), filtered using 40âµm filters and live cells were counted using Guava Muse cell counter (Cytek) and the Muse Count and Viability Kit (Cytek). Equal numbers (10,000) of live colonocytes per condition in a 20âµl Matrigel (70% Matrigel) dome were plated in a prewarmed 48-well plate and cultured for 6â7 days and were longitudinally imaged using the Incucyte S3 Live-Cell Analysis Instrument (Sartorius). Cells from DMSO-treated or GW3965-treated (1âµM) conditions were cultured in respective WENR stem cell medium for first 3âdays, followed by differentiating ENR medium with subsequent medium change every 2âdays. Organoid numbers on day 4 counted using the Incucyte Organoid Analysis Software Module were used to estimate the organoid plating efficiency (that is, the number of organoids/10,000âÃâ100). Furthermore, owing to non-budding nature of the colon organoids, the total organoid area was estimated using the Incucyte organoid module. Colon organoids were collected at day 6/7 after single-cell seeding for gene expression analysis using qPCR.
Image analysis protocol using the Incucyte Organoid Analysis Software Module
A set of representative images was chosen to ensure comprehensive coverage of organoid structures. The background to cell ratio was adjusted to accurately define the boundaries of organoid objects. Subsequently, the bright-field mask was evaluated, and filter parameters were refined accordingly. The refined bright-field mask was then applied to the image set, and a verification was conducted to ensure effective masking across all representative images. The edge split parameter was adjusted to delineate between individual organoid objects and refine segmentation. The bright-field mask was re-evaluated, and filter parameters were refined to ensure optimal delineation. After confirming that the parameters appropriately masked all organoids in the representative images, the Launch Wizard analysis was completed by selecting the scan times and wells to be analysed. This final step ensured a systematic and thorough analysis of organoid structures in the selected images.
Salivary gland organoids
Salivary gland organoids were cultured according to the protocol published before34. Salivary glands from WT (C57BL/6J) mice were collected in 2,880âμl of RPMI (Sigma-Aldrich) supplemented with 2âmM l-glutamine, 1à penicillinâstreptomycin (Thermo Fisher Scientific), 5% FCS and 0.1âmgâmlâ1 DNase I (Roche). The salivary glands were minced with scissors into small pieces followed by addition of 120âμl of 25âmgâmlâ1 of collagenase D (Sigma-Aldrich) to the 2,880âμl of RPMI. Tissues were incubated at 37â°C with gentle agitation for 40âmin. Digestion was stopped by adding 30âμl of 0.5âM EDTA (Invitrogen). Cell suspensions were filtered using 100âµm cell strainers (Corning), centrifuged at 300g for 8âmin at 4â°C. The cell pellet was further digested with TrypLE Express (Gibco) with 1,200âUâmlâ1 of DNase I (Roche) at 37â°C for 10âmin. Cells were then washed, resuspended in advanced DMEM/F12 (Gibco) with 1à HEPES (Thermo Fisher Scientific) and then plated for organoids. Cells in basic culture medium were mixed with Matrigel (Corning) at a ratio of 30 (medium):70 (Matrigel) and 20âμl of mixture was plated carefully on to the centre of prewarmed flat bottom 48-well cell culture plate. The plate was placed at 37â°C and allowed to solidify for 15âmin before 200âμl of organoid medium was added to each well. The basic culture medium contained advanced DMEM/F12, 1à penicillinâstreptomycin, 1à GlutaMAX (Thermo Fisher Scientific), 10âmM HEPES (Thermo Fisher Scientific), 1à B27 (Life Technologies), 1à N2 (Life Technologies) and 1âmM N-acetylcysteine (Sigma-Aldrich), and was supplemented with 0.2âµgâmlâ1 Primocin (InVivoGen), 100ângâmlâ1 of murine noggin (Peprotech), 100ângâmlâ1 recombinant murine R-spondin (R&D), 5ânM FGF1 (Peprotech), 1ânM FGF7 (Peprotech), 5ânM NRG1 (R&D) and 0.5âµM A83-01 (Merck). The medium was changed every 3âdays and the cultures were maintained at 37â°C in fully humidified chambers containing 5% CO2. For first 7âdays of culture, 10âµM Y-27632 (Sigma-Aldrich) was added in the medium. On day 7, the salivary glands were washed to clean the Matrigel and replated without breaking the organoids. On day 7, while replating, DMSO or GW3965 (1 µM, Sigma-Aldrich) was added to the organoid medium without further addition of Y-27632. Then, 3âdays after (day 10), when the medium was replenished, DAPT (5âµM, Tocris) was added to the culture medium. Organoids were imaged using the Nikon SMZ25 microscope with the NIS Elements software and collected for gene expression analysis between day 11 and day 13. For quantification of organoid area, z stacks of whole-well images were converted to a focused EDF document and, using Affinity Photo 2, the contrast and gamma ratio were adjusted for each image, cropped in oval mode to remove edges but take all organoids. Segmentation analysis was performed using OrganoSeg software53 according to the instructions and the organoid area was measured for each organoid and averaged per well from four different experiments.
Single-cell sorting for organoid co-culture
To obtain single cells, isolated SI crypts were digested with TrypLE Express (Gibco) with 1,200âUâmlâ1 of DNase I (Roche) at 32â°C for 90âs. Cells were then washed and stained with the following antibodies: CD31-PE (BioLegend, Mec13.3), CD45-PE (eBioscience, 30-F11), Ter119-PE (BioLegend, Ter119), EPCAM-APC (eBioscience, G8.8) and CD24-Pacific Blue (BioLegend, M1/69) at 1:500 dilutions by incubating for 30âmin on ice. After staining, cells were washed in SMEM (Sigma-Aldrich), filtered using 70âµm filter and resuspended in SMEM supplemented with 7-AAD (BD Bioscience, 1:500). Cells were sorted using the FACS ARIA II (BD Biosciences) system. Sorting strategies were as follows: ISCs, Lgr5eGFPhighEPCAM+CD24med/âCD31âTer119âCD45â7-AADâ; Paneth cells, CD24highsidescatterhighLgr5eGFPâEPCAM+CD31âTer119âCD45â7-AADâ. For organoid co-culture, ISCs were sorted from Lgr5-eGFP-creERT2 mice and PCs were sorted from non-Lgr5 WT mice. Equal numbers (~5,000 each) of ISCs and PCs were co-cultured using standard ENR medium further supplemented with additional 500ângâmlâ1 R-spondin (to have a final concentration of 1,000ângâmlâ1), 100ângâmlâ1 WNT3A (R&D Systems) and 10âµM jagged-1 peptide (Anaspec) for first 4âdays together with 10âµM Y-27632. After 4âdays, the medium was changed to regular ENR medium. For in vitro stimulation, LXR agonist (1âµM GW3965) was added in the medium for the entire duration of the organoid culture. DMSO was used as vehicle. Crypt domain budding was quantified on day 8 after starting the co-culture.
Cell isolation and cell sorting from mouse SI
Cell isolation from the mouse intestine performed using previously published protocol5 with a few modifications. In brief, dissected SIs were flushed with cold PBS to remove the intestinal content and mucus, then cut open longitudinally and cut into ~1âcm pieces. To isolate the intestinal epithelial cells (IECs) and the intraepithelial lymphocytes (IELs), intestinal pieces were incubated in HBSS (HyClone) supplemented with 5% FCS, 15âmM HEPES (Thermo Fisher Scientific), 5âmM EDTA (Life technologies) and 1âmM DTT for 30âmin at 37â°C under agitation at 800ârpm. The supernatant (called IEL fraction) containing IECs and intraepithelial lymphocytes was filtered through 100âµm cell strainers, centrifuged at 500g for 5âmin at 4â°C and was enriched using a 44%/67% Percoll gradient and washed for FACS analysis. To further isolate the cells from the LP, the intestinal pieces were washed with PBS supplemented with 5% FCS and 1âmM EDTA for 15âmin at 37â°C under agitation at 800ârpm. Next, the tissue pieces were digested in HBSS supplemented with 0.15âmgâmlâ1 liberase TL (Roche) or 0.5âmgâmlâ1 collagenase D (Sigma-Aldrich) and 0.1âmgâmlâ1 DNase I (Roche) at 37â°C for 45âmin under agitation at 800ârpm. The digested tissues were filtered through 100âµm cell strainers and were washed for FACS analysis (called LP fraction). Single-cell suspensions were blocked with Fc-blocking solution (1:1,000, eBioscience) and stained with the antibody mix at 4â°C for 15âmin. To examine immune cell subsets, acquired single SI cells were stained with an antibody cocktail of EPCAM-FITC (BioLegend, G8.8), CD19-FITC (BioLegend, 6D5), CD45-BV650 (BioLegend, 104), CD90-APC-Cy7 (BioLegend, 30-H12), CD64-BV421 (BioLegend, X54-5/7.1), CD11b-BV786 (Invitrogen, M1/70), CD11c-PE-Cy7 (BioLegend, N418), MHC-II (BD-Biosciences, 1A8), Ly6G-PE (BD-Biosciences, 1A8), Ly6C-PcP5.5 (Invitrogen, HK1.4), CD3-AF700 (BD-Biosciences, 500A1), CD103-APC (Invitrogen, 2E7) at 1:200 dilution for 15âmin followed by flow cytometry analysis. To sort SI cells, the following antibodies were used for the staining: EPCAM-FITC (BioLegend, G8.8, 1:200), CD45-PeCy7 (BioLegend, 104, 1:200), Ter119-Pacific Blue (PB) (BioLegend, Ter119, 1:200). Cells were sorted using the SONY SH800S cell sorter. Sorting strategies were as follows: epithelial cells (IECs), EPCAM+CD45âTer119âDAPIâ; intraepithelial immune, CD45+EPCAMâTer119âDAPIâ from the IEL fraction; LP immune, CD45+EPCAMâTer119âDAPIâ; DN, CD45âEPCAMâTer119âDAPIâ from LP fraction. Sorted cells were collected in RPMI (Sigma-Aldrich) medium supplemented with 5% FCS, centrifuged and resuspended in Trizol for RNA extraction. Flow cytometry data and dot plots were analysed and prepared using FlowJo (v.10).
Sample preparation for scRNA-seq in SI tissue and organoids
For scRNA-seq from the SI, WT C57BL/6J mice were fed with either a standard (nâ=â3) or GW3965 diet (nâ=â3) for 10 days. Next, cell suspensions were prepared and the respective fractions (that is, IEL and LP) were pooled before sorting IECs, immune and DN cells as described above. For scRNA-seq analysis of SI organoids, organoids were established using SI crypts from WT C57BL/6J mice (nâ=â2) and grown with NR culture medium and were treated with either DMSO or 1âµM GW3965 for 5 days with medium change every 48âh. On day 5, organoids were removed from Matrigel, cleaned with cold PBS and digested with TrypLE Express (Gibco) with 1,200âUâmlâ1 of DNase I (Roche) at 32â°C for 5âmin. After digestion, cells were washed and filtered using 70âµm filter. Single-cell suspensions were blocked with Fc-blocking solution (eBioscience, 1:1,000) and stained with the antibody mix (1:200) at 4â°C for 15âmin. The following antibodies were used for the staining: EPCAM-FITC (BioLegend, G8.8), CD45-PeCy7 (BioLegend, 104), Ter119-Pacific Blue (BioLegend, Ter119). EPCAM+CD45âTer119âDAPIâ epithelial cells (IECs) were sorted using the SONY SH800S cell sorter. Approximately 30,000 cells from each condition were loaded onto the 10x chip and the samples were processed using the Chromium Next GEM single cell 3â² reagents kit v3.1 (dual index).
scRNA-seq analysis
Libraries prepared for scRNA-seq were sequenced by Novogene at a sequencing depth of 300âmillion reads per sample using paired-end 150âbp reads on the NovaSeq 6000 sequencer and using the v1.5 reagent kit. Raw sequences were quantified and annotated using CellRanger (10x Genomics) v.6.0.2 (organoids) or v.6.1.2 (SI). Annotated gene counts were processed with Seurat54 v.3.6.3 (organoids) or v.4.1.1 (SI tissue). Cells were filtered to include only cells with >500 (organoids) or 400 (SI) unique genes expressed, >5% ribosomal RNA and <10% mitochondrial RNA. SI samples were also filtered for cells expressing <25% Hb genes. Only genes expressed at >3 counts across all cells were included in the dataset. Mitochondrial genes were also excluded from further analyses. Doublet cells were excluded using DoubletFinder55.
For the organoid dataset, integration of the two samples was performed using the first 30 dimensions of CCA in Seurat. Clustering was performed using FindNeighbors (dims = 1:30, k.param = 200, prune.SNNâ=â1/15) and FindClusters (original Louvain algorithm at resolution 0.5). Differential expression was analysed using FindAllMarkers (Wilcoxon rank-sum test, log[fold change]â>â0.2, Pâ<â0.01) for cluster markers (subsampling 50 cells per cluster, difference in percentageâ>â0.2) and diet groups within clusters (subsampling 150 cells).
For the SI dataset, a primary UMAP was performed using all samples (based on the first 20 components of a principal component analysis (PCA) based on the 2,000 most highly variable genes). The two epithelial samples were integrated as for the organoid dataset and clustering was performed (FindNeighbors parameters: dimsâ=â1:30, k.paramâ=â200, prune.SNNâ=â1/15). The four immune and double negative samples were integrated as two samples, considering the two samples from the same animals as one, and clustering was performed as for the epithelial samples. Clusters were annotated manually using differential gene expression (FindAllMarkers as above). Differential expression analysis was performed between the standard-food and GW3965 samples for each sorting fraction separately using FindMarkers (Wilcoxon rank-sum test, subsampling 500 cells per group). The expression of inflammatory response genes was calculated using the Seurat function AddModuleScore using the genes included in GO term inflammatory response (GO: 0006954).
Tissue processing and spatial transcriptomics of SI and colon
SI tissues from standard- and GW3965-diet fed mice at 0 and 3âd.p.i. were used for spatial transcriptomics. Colonic tissues from standard- and GW3965-diet fed mice at day 22 and day 43 of AOMâDSS treatment and untreated mice fed with standard diet only (day 0) were processed for spatial transcriptomics. In brief, SI and colon were cleaned from adipose tissue and cut longitudinally to remove the luminal content by washing in ice-cold PBS. Starting from the most distal portion, that is, ileum for SI and rectum for colon, the tissues were rolled, resulting in a Swiss roll with the most distal part in the centre and the proximal part in the outer portion of the roll. The Swiss rolls were snap-frozen for 1âmin in a liquid nitrogen-cooled isopentane bath within a plastic tissue cassette. The frozen tissues were then embedded in optimal cutting temperature compound (OCT, Sakura Tissue-TEK) on dry ice for slow cooling and stored at â80â°C. Tissue sections (thickness, 10âµm) were cut using a pre-cooled cryostat and the tissue sections were transferred onto the oligo-barcoded capture areas (6.5âmm2) on the 10x Visium Genomics slide and stored at â80â°C until further processing. The Visium spatial transcriptomics protocol was performed according to previously published studies19 and the manufacturerâs instructions (10x Genomics, Visium Spatial Transcriptomic). H&E stained images were captured using a Leica DM5500 B microscope (Leica Microsystems) at 5X magnification. The Leica Application Suite X (LAS X) was used to acquire tile scans of the entire array and merge images. Sequence libraries were then processed according to the manufacturerâs instructions (10x Genomics, Visium Spatial Transcriptomic). After the second cDNA strand synthesis, cDNA was quantified using the RT-qPCR ABI 7500 Fast RealTime PCR System and analysed using ABI 7500 v.2.3.
Visium sequencing and data processing
Visium libraries were sequenced on the NovaSeq S1 flow cell (Illumina), at a depth of around 200âmillion reads per sample, with 28 bases from read 1 and 120 bases from read 2. FASTQ files were processed with SpaceRanger (10x Genomics) v.1.1.0 (colon) or v.1.2.0 (SI) mapped to the pre-built mm10 reference genome (GRCm38).
Spatial transcriptomic analyses of SI tissues
The SI datasets was analysed using Seurat v.4.1.1 in R v.4.0.5 as follows. SpaceRanger output was imported and spots with fewer than 20 unique expressed genes were removed, as were genes expressed in fewer than 5 spots. PCA was performed using the 4,000 most highly variable genes. Harmony56 was used to integrate the four samples. Graph construction was performed with the RcppHNSW57 package using the first 50 dimensions in harmony, kâ=â20 and cosine distance, followed by Louvain clustering using the igraph58 package. Differential expression between clusters was computed using Seurat function FindAllMarkers, with genes considered to be significant at FDRâ<â0.05 and log2[FC]â>â0.25. Differential expression between standard diet samples at 0âd.p.i. and 3âd.p.i. was calculated using the Seurat function FindMarkers. Changes in Areg expression between conditions were analysed by identifying Areg+ spots (read countsâ>â0) and performing differential expression between Areg+ spots in standard-diet and GW3965 samples at 3âd.p.i., rerunning the analysis for each cluster separately. Non-negative matrix factorization was performed using the cNMF package in Python (v.3.9)59.
Spatial quantification of single cells during CRC progression
Visium sequencing data were processed using SpaceRanger software v.1.1. Six colonic sections were used for analysis, spanning from healthy control (that is, 43 days of standard diet without any AOMâDSS treatment, defined as day 0), AOMâDSS-induced CRC on standard diet at two timepoints (day 22 and day 43) and AOMâDSS-induced CRC on a diet containing GW3965 at two timepoints (GW day 22 and GW day 43). We also used a public single-cell dataset (GSE148794)49 containing the major immune, stromal and epithelial cells during the time course of DSS colitis, both at steady state and during inflammation. Single-cell and spatial transcriptomics data were analysed using Seurat54. Genes related to mitochondrial and Malat1 as well as those detected in at least 5 cells were omitted from the downstream analysis. The top 4,000 highly variable genes were used for denoising using PCA (50 PCs), which in turn was used for visualization using UMAP (https://arxiv.org/abs/1802.03426), k-nearest neighbour graph construction on cosine distance and finally clustering using Louvain (resolutionâ=â1). Cell clusters were annotated based on differentially expressed markers genes49. Projection of the aforementioned single-cell clusters onto the CRC Visium dataset was done using weighted non-negative least squares implemented in the SCDC package60, a robust and fast method for cell type deconvolution61. Cell type abundances were compared pair-wise between conditions and their respective control (either day 0 or day 22/day 43) using Wilcoxon tests. Alterations in cellâcell co-detection in spatial transcriptomics spots was quantified and compared between GW3965 and standard diets, using the approach previously described62. Significant alterations were projected back to the single-cell UMAP embeddings for illustration.
Bulk RNA-seq analysis of AOMâDSS tumour kinetics
cDNA libraries were constructed and sequenced by Novogene at a sequencing depth of 20âmillion reads per sample using paired-end 150âbp reads on the NovaSeq 6000 sequencer and the v1.0 reagent kit. Raw sequences were quantified and annotated using Kallisto63 and GRCm38 (mm10) cDNA assembly64. Resulting gene counts were used for the following analyses, which were performed in R v.3.6.3 (R Core Team, 2020, https://www.R-project.org/). Owing to important differences in RNA degradation and the ribosomal RNA percentage between samples, analyses included corrections for the percentage of ribosomal RNA as described below. DEGs were identified with edgeR65, according to the linear model yâ~âdietâÃâtimeâÃâpercentage of ribosomal RNA. Genes were included in the module analysis if FDRâ<â0.05 in a likelihood ratio test including the diet and diet:interaction terms of the model (H0: coefficients for all included terms equal 0). For all DEGs, log[FC] values were calculated for each group by combining log[FC] values from the likelihood ratio test for each relevant term (for example, log[FC]d70-GW3965â=â0 + log[FC]d70â+âlog[FC]GW3965â+âlog[FC]d70:GW39765). These log[FC] estimates were scaled by the mean and s.d., and scaled log[FC] values of DEGs were subsequently clustered into modules using hierarchical clustering (Euclidean distance, Wardâs method, tree cut at height 20). Over-represented KEGG pathways66 and GO biological process terms22 were identified using Enrichr (FDRâ<â0.05). To evaluate intragroup variability, module expression for each sample was calculated as the mean expression of all scaled log-transformed normalized counts (given by the edgeR cpm function, corrected for the percentage of ribosomal RNA using the limma function removeBatchEffect after log-transformation).
RNA in situ hybridization (RNA scope)
RNA in situ hybridization was used to detect the expression of Lgr5, Abca1 and Areg (Advanced Cell Diagnostics), in intestinal tissues at steady state and after irradiation. Formalin-fixed paraffin-embedded SI Swiss rolls were sectioned at 5âμm depth and were captured in MiliQ water using Superfrost gold slides. The RNAscope Multiplex Fluorescent v2 Assay (Advanced Cell Diagnostics) was used according to the manufacturerâs protocol. In brief, the paraffin sections were baked for 1âh at 60â°C in the HybEZ Oven, then deparaffinized in xylene and washed in 100% ethanol. RNAscope hydrogen peroxide was used to block endogenous peroxidase activity by incubating slides at room temperature for 10âmin and RNAscope 1à target retrieval was performed at 100â°C for 15âmin. The slides were then incubated with RNAscope Protease Plus for 30âmin at 40â°C in the HybEZ Oven. Lgr5 (C2) and Areg (C3) probes were diluted in Abca1 (C1) probe at a 1:50 ratio, the mix was added to the slides and baked for 2âh at 40â°C in the HybEZ Oven. The hybridization was done for AMP1, AMP2 and AMP3 according to the manufacturerâs protocol. The Abca1 signal was developed using TSA Vivid Fluorophore kit 570, Lgr5 using Fluorophore kit 520 and Areg using Fluorophore kit 650 (Tocris). Finally, the counterstaining with DAPI was performed for 30âs at room temperature and the slides were mounted using ProLong Gold Antifade mounting medium (ProLong Antifade reagent; Invitrogen) and scanned with the Zeiss LSM 880 with Airyscan confocal laser-scanning microscope (Carl Zeiss) using a Ã20 air objective. Images were processed using Fiji/ImageJ (NIH).
Immunohistochemistry on paraffin embedded murine tissues
Tissues were fixed in 10% buffered formalin, paraffin embedded and sectioned (5âµm). For immunohistochemical analysis, the sections were deparaffinized in xylene and then rehydrated with graded alcohols. Endogenous peroxidase was blocked using 3% H2O2 (Scharlau) in methanol for 1âh. Antigen retrieval was performed using either 1âmM EDTA buffer (pHâ8.0) or 10âmM Na-citrate buffer (pHâ6.0) at 121â°C for 20âmin using 2100 Antigen Retriever. The slides were then washed with PBS and blocked using BLOXALL solution (BLOXALL Blocking solution; Vector Labs) for 2âh at room temperature. The sections were incubated overnight at 4â°C with primary antibodies diluted in PBS. Antibodies used were as follows: rat anti-BrdU 1:300 (Abcam), rabbit anti-cleaved caspase 3 1:200 (Cell Signaling Technology), rabbit anti-CYP27A1 1:200 (Abcam), mouse anti-AREG 1:500 (Santa Cruz) and rat anti-B220 1:200 (BioLegend). The slides were washed four times with PBS and incubated with biotinylated goat anti-rabbit (Vector Labs, 1:300), biotinylated goat anti-rat (Vector Labs, 1:300) or biotinylated goat anti-mouse (Vector Labs, 1:300) for 1âh at room temperature. The slides were washed four times in PBS and were incubated with the VECTASTAIN Elite ABC HRP Kit (Vector Labs) for 30âmin. Staining was developed using DAB staining (DAB peroxidase staining kit, Vector Labs). The sections were counterstained with haematoxylin (Harris hematoxylin, Leica) and washed in tap water, dehydrated in increasing grades of alcohol and then with xylene and dried. The slides were mounted using permanent mounting medium (VectaMount, Vector labs). The slides were scanned using the Hamamatsu NanoZoomer Slide Scanner. Sectioning, H&E staining and DSS colitis and AOMâDSS tumour grading was performed in a blinded manner by the FENO (Morphological Phenotype Analysis) facility (Karolinska Institutet).
Quantification of immunohistochemistry staining
Quantification of cCASP3+ and BrdU+ cells in the SI crypts was performed in a blinded manner using QuPath software. To isolate and quantify the distal SI, the first 10âcm of the proximal SI (starting from the pylorus) was removed and the rest of the distal SI was Swiss-rolled. Crypts (approximately 100 crypts per section) were delineated manually using the polygon or brush tool in QuPath. Crypts were selected if sectioned whole length (that is, not transversally cut). The numbers of DAB+ cells per crypt were quantified automatically using the âpositive cell detectionâ command in QuPath. The percentage of surviving crypts was calculated as the percentage of crypts with more than or equal to 10 BrdU+ cells out of all of the crypts quantified. The average number of cCASP3+ or BrdU+Â cells per crypt was calculated as the average number of DAB+ cells in all of the crypts analysed in each section. The normalized percentage of surviving crypts (where indicated), was calculated by dividing the percentage of surviving crypts in each experiment by the average of the percentage of surviving crypts in the control group of each experiment.
For quantification of LP CYP27A1+ cells in the crypt niche, an average of 48 intact crypts within Swiss rolls stained for CYP27A1+ at different âd.p.i. were first identified using NDP.View 2 (NanoZoomer Digital Pathology) software. DAB+ cells in the epithelial crypt base, transit-amplifying zone and LP surrounding each crypt were manually counted.
Quantification of total CYP27A1 staining in SI and colon Swiss-rolls was quantified using ImageJ. In brief, scanned sections were converted to black and white using the âmake binaryâ command in ImageJ and the total mean grey value of the stained area was calculated. In the colour threshold tab, hue, saturation and brightness were adjusted to highlight only the DAB+ signal. Using the make binary command, the mean grey value of the DAB intensity was then calculated. The CYP27A1 intensity was calculated as the ratio of the mean grey value of the DAB intensity by the mean grey value of the total stained area (that is, area of the total Swiss-roll). The normalized CYP27A1 intensity was calculated by dividing the intensity of each timepoint (for example, post-irradiation) by the average of the intensity of the untreated control (for example, day 0).
AREG quantification in the crypt and villus LP of the distal SI was performed in a blinded manner. For crypt AREG quantification, around 10 high-powered field (Ã40) images per mouse were exported and analysed using QuPath. In each image, crypts (4â15 crypts per field) were delineated manually using the polygon or brush tool as described previously. Overall around 100â125 crypts per mouse were quantified. Using the automated quantification tool, mean DAB intensity and the crypt area were quantified for each crypt. Mean DAB intensity for each crypt was normalized to its area, which was then used to calculate the average DAB intensity (AREG staining) per mouse. For AREG staining in villus LP, 10X images of the complete distal SI was exported and analysed using QuPath. In brief, the crypt regions were selected out and the analysis was focused only on the villus region. Using the cell detection tool, the mean DAB intensity and the area of individual selected cells was quantified. Using this, we quantified around 300â1,500 cells in the villus region/mouse. The mean DAB intensity for each cell in the villus LP was normalized to its area, which was then used to calculate the average DAB intensity (AREG staining) per mouse.
Quantification of B220+ B cell follicles in AOMâDSS tumour samples was done blinded using QuPath software. In brief, the anti-B220 stained follicles irrespective of their sizes or staining intensity were selected and outlined using a polygon or brush tool in QuPath. The number of annotated B220+ follicles was counted and plotted.
Immunofluorescence of mouse paraffin tissues
Tissue fixation, paraffin embedding, sectioning and staining protocols were performed as described above for immunohistochemistry with the following changes. For primary antibody detection, slides were incubated in PBS with rabbit anti-OLFM4 (1:300, Cell Signalling), rat anti-BrdU (1:300, Abcam) and wheat germ agglutinin (WGA) AF555 (1:5,000, Invitrogen) overnight at 4â°C in darkness. After primary antibody incubation, the slides were washed with PBS and stained with goat anti-rabbit AF488 (1:500, Invitrogen), donkey anti-rat AF647 (1:500, Jackson ImmunoResearch) for 90âmin at room temperature in darkness. After secondary antibody staining, the slides were washed with PBS and nuclei were stained with DAPI (1âµgâmlâ1) for 30âmin and subsequently washed with PBS. Mounting was performed using Vibrance Antifade Mounting Medium (Vectashield). Fluorescent-stained slides were imaged using the LSM880 confocal microscope (Zeiss). Quantification of BrdU+Olfm4+ and Olfm4+ cells from 20â30 ileal crypts per mouse was performed using the Cell Counter plugin in Fiji/ImageJ (NIH). WGA staining was used to differentiate the crypt niche and the TA zone.
Immunoblotting
SI organoids were grown in ENR or NR medium supplemented with DMSO, GW3965 (1âμM), with the medium changed every second day. On day 5, organoids were lysed in RIPA buffer with 1à protease inhibitor cocktail (Roche) and 1à PhosStop (Roche) phosphatase inhibitors. Cells were sonicated, centrifuged for 5âmin at 10,000ârpm, and the cleared lysates were measured using the DC Protein Assay (Bio-Rad). The samples were run on 4â12% Bis-Tris protein gels (Thermo Fisher Scientific) with 20ângâμlâ1 protein loaded per well and the Precision Plus Protein All Blue Standards (Bio-Rad, 1610373EDU) as a molecular mass marker. Gels were blotted onto PVDF membranes, followed by blocking in 5% non-fat dry milk and incubated overnight at 4â°C with the following primary antibodies: rabbit polyclonal ERK1/2 (Cell Signaling Technology; 1:1,000), rabbit polyclonal phospho-ERK1/2 (Cell Signaling Technology; 1:1,000), and mouse monoclonal Vinculin (Sigma-Aldrich, V9131, 1:1,000). HRP-conjugated anti-rabbit (Sigma-Aldrich; 1:5,000) or anti-mouse (Cell Signaling Technology; 1:1,000) were used as secondary antibodies for 1âh at room temperature. Signal was detected using the Pierce SuperSignal West Pico Plus reagent (Thermo Fisher Scientific) and visualized on the BioRad luminescence detector. Densitometry was performed using the software Image Lab.
RNA extraction and RTâqPCR
Tissue biopsies collected for RNA extraction were preserved in RNAlater (Invitrogen) at â20â°C. Tissues were transferred to RLT Plus buffer (Qiagen)â+â2% β-mercaptoethanol and lysed by bead-beating (Precellys) and stored at â80â°C till RNA extraction. Intestinal crypts and organoid cell suspensions were incubated in RLT Plus buffer (Qiagen)â+â2% β-mercaptoethanol and stored at â80â°C before RNA extraction. RNA isolation was performed using the RNAeasy Mini Kit (Qiagen) according to the manufacturer instructions and was quantified using the Nanodrop. For RNA extraction from cells sorted directly into Trizol (Life technologies) or resuspended in Trizol after sorting, sorted cells were mixed well in Trizol and incubated for 5âmin at room temperature and stored at â80â°C until further processing. For RNA extraction, samples were thawed at room temperature and an appropriate amount of chloroform (Fisher Scientific) was added and mixed by vortexing it well followed by 2â3âmin incubation at room temperature and then centrifuging at 12,000g for 15âmin at 4â°C. The aqueous phase containing the RNA was carefully transferred to new Eppendorf tubes and 1âµl of Glycoblue (Thermo Fisher Scientific) was added to each of the samples, which were then mixed by flicking; an appropriate amount to isopropanol was then added to each sample and the samples were mixed well. The samples were then incubated for 1âh at â80â°C followed by centrifuging at 12,000g for 20âmin at 4â°C. The samples were then washed in 70% cold ethanol by centrifuging at 12,000g for 15âmin at 4â°C. Excess ethanol was removed and the samples were air dried until all of the residual ethanol was removed. The samples were resuspended in an appropriate amount of nuclease-free water (Life Technologies) and were quantified using the Nanodrop. Reverse transcription was performed using iScript RT Supermix (Bio-Rad). Gene expression was analysed by qPCR using the iTaq Universal SYBR Green Supermix (Bio-Rad) and log2-transformed fold changes were calculated relative to either Hprt (when using tissues) or B2m (when using cells) or Actb (when indicated) mRNA using the \({2}^{-\Delta \Delta {C}_{{\rm{t}}}}\) method67. For mouse Cyp27a1 expression, primer pair 1 was used when expression was analysed in WT mice, and primer pair 2 was used when expression was analysed in Cyp27a1â/â or control littermates. Primers used for qPCR were as follows: b2m, ACCGTCTACTGGGATCGAGA, TGCTATTTCTTTCTGCGTGCAT; Hprt, TCAGTCAACGGGGGACATAAA, GGGGCTGTACTGCTTAACCAG; Actb, CACTGTCGAGTCGCGTCC, GTCATCCATGGCGAACTGGT; Abca1, TGGGCTCCTCCCTGTTTTTG, TCTGAGAAACACTGTCCTCCTTTT; Abcg1, AGGCAGACGAGAGATGGTCA, AAGAACATGACAGGCGGGTT; Olfm4, TGCTCCTGGAAGCTGTAGTCA, TGTATTCAAAGGTGCCACCCA; Areg, CAGTGCACCTTTGGAAACGA, ATGTCATTTCCGGTGTGGCT; Ereg, TGCTTTGTCTAGGTTCCCACC, CGGGGATCGTCTTCCATCTG; Tgfa, CAAACACTGTGAGTGGTGCC, GGGATCTTCAGACCACTGTCTC; Egf, AGGATCCTGACCCCGAACTT, ACAGCCGTGATTCTGAGTGG; Egfr, CGCCAACTGTACCTATGGATGT, GGGCCACCACCACTATGAAG; Wnt3, TGGAACTGTACCACCATAGATGAC, ACACCAGCCGAGGCGATG; Jag1, GAGCCAAGGTGTGCGG, GCGGGACTGATACTCCTTGA; Jag2, GCCTCGTCGTCATTCCCTTT, AGCTCCTCATCTGGAGTGGT; Dll1, GCGACTGAGGTGTAAGATGGA, GCAGCATTCATCGGGGCTAT; Hes1, GAAAAATTCCTCCTCCCCGGT, GGCTTTGATGACTTTCTGTGCT; Cyp27a1 (pair 1), CCCACTCTTGGAGCAAGTGA, CCATTGCTCTCCTTGTGCGA; Cyp27a1 (pair 2), CCCAAGAATACACAGTTTG, GCCTCTTTCTTCCTCAGC; Il33, GGGCTCACTGCAGGAAAGTA, TGGGATCTTCTTATTTTGCAAGGC; Clu, ATTCTCCGGCATTCTCTGGG, CCTTGGAATCTGGAGTCCGGT; Msln, GCCTAGTAGACACTACTGCAGAC, AGCAGTAGGAAGCTTCGGC, Yap1, ATTTCGGCAGGCAATACGGA, CACTGCATTCGGAGTCCCTC; Wnt9b, CCAGAGAGGCTTTAAGGAGACG, GGGGAGTCGTCACAAGTACA.
For gene expression analysis in human biopsies, the following Taqman probes from Thermo Fisher Scientific were used: HPRT1: Hs02800695_m1_4453320; CYP27A1: Hs01017992_g1_4448892; ABCA1: Hs01059137_m1_4448892.
Cohort of patients with ulcerative colitis
Paired endoscopic biopsy specimens were obtained from the ascending colon and sigma/rectum from patients with IBD or suspicion of intestinal disease. One of the paired biopsies was used for histological assessments and the other for RNA extraction. Human studies were approved by the local ethical committee (EthikKommission der Ãrztekammer Hamburg PV4444).
Human CRC tissue microarray
Patients
Samples from patients with CRC from the Coloproctology Department, ClÃnica Las Condes, were included between 2015 and 2017. All of the patients signed informed consent forms approved by the institution (Cómite de ética de la investigación de CLC, O22019AA) and the procedures were performed according to human experimental and clinical guidelines. Patients undergoing surgery for tumour resection had to be older than 18 years old and not have received chemotherapy or neoadjuvant therapy before total or partial colectomy. Tumour staging was classified according to the TNM classification (The Union for International Cancer Control; UICC). Immediately after surgery, samples of fresh tumour and healthy intestinal mucosa (at least 10âcm away from tumour) were macroscopically selected. Biopsy-size samples of tumour and healthy tissue were fixed in 2% paraformaldehyde and paraffin embedded for a tissue microarray (TMA) construction and immunohistochemistry analysis.
TMA generation
For TMA generation, TMAs were assembled from formalin-fixed paraffin-embedded tissues using a 0.6-mm-diameter punch (Beecher Instruments). The arrays encompass 14 tissue cores from colonic tumours and healthy tissue derived from 11 patients. Moreover, two cores from kidney tissue were used (for orientation purposes). Using a tape-transfer system (Instrumedics), 2âμm sections were transferred to glass slides and analysed using immunohistochemistry (IHC).
IHC
Conventional IHC on TMA sections was performed on 2âμm sections using antibodies against CYP27A1 (Abcam, dilution 1:200). Immunohistochemistry was performed using the R.T.U. VECTASTAIN Kit (Vector Labs), according to the manufacturerâs instructions. Sections were deparaffinized and rehydrated with deionized water. The sections were then heated in EDTA buffer, pHâ8.0, for 20âmin, and cooled for 10âmin before immunostaining. All of the samples were blocked by exposure to R.T.U. normal horse serum for 15âmin, then incubated in the following sequential order: primary antibody for 1âh, 3% H2O2 (blockade of endogenous peroxidases) for 15âmin, R.T.U biotinylated universal antibody anti/rabbit/mouse IgG for 30âmin, R.T.U. ABC reagent, 3â²3-diaminobenzidine as a chromogen for 5âmin and finally counterstained with haematoxylin for 5âmin. The above was carried out at room temperature and, between incubations, the sections were washed with Tris-buffered saline. Coverslips were placed using the Tissue-Tek SCA (Sakura Finetek). Images were captured using the Aperio ScanScope equipment, analysed by the Aperio ImageScope Software and evaluated using the Positive Pixel Count 9 algorithm. The proportion of positive pixels with respect to the total pixels (positive and negative) per area was evaluated in crypts and tumour region, excluding stroma (positivity/area).
Indirect immunofluorescence
Paraffin histological sections derived from a primary CRC tumour and healthy tissue were evaluated for the co-expression of CYP27A1 and Vimentin by immunofluorescence. In brief, the sections were subjected to deparaffinization (NeoClear, Merck), then rehydrated with a battery of alcohols from absolute ethanol to 70% ethanol. The antigen retrieval was performed with EDTA buffer (pHâ8.0). The sections then were incubated with 100âmM glycine and 2% BSA (SigmaâAldrich)â+â1% normal donkey serum in 1Ã PBS (SigmaâAldrich) (for autofluorescence and non-specific protein blocking, respectively). The sections were incubated at room temperature for 1âh with the following primary antibodies: rabbit anti-CYP27A1 (1:200, Abcam) in conjunction with mouse anti-vimentin (1:1,000, Abcam). After a rinse with PBS, the tissue sections were incubated for 1âh at room temperature with the following secondary antibodies (Invitrogen): goat anti-rabbit IgG conjugated with Alexa Fluor 488 (1:500) and goat anti-mouse IgG conjugated with Alexa Fluor 546 (1:200). Hoechst 33342 (1:500) was used as a nuclear counterstain. Finally, the slides were covered with a coverslip plus mounting solution (Dako, Agilent Technologies). The slides were visualized using the C2+ confocal microscope with the Ã20 objective (Nikon Instruments).
Bulk and single-cell RNA-seq reanalyses of published datasets
Re-analysis of deposited scRNA-seq datasets
The following publicly available datasets were downloaded from the Gene Expression Omnibus (GEO): GSE117783 (ref. 4), scRNA-seq data from both normal and irradiated SI crypts. The standard Seurat v.3.1.3 protocol was used to analyse these datasets. Cells with a number of expressed genes of <200 and >10,000 were first filtered out. Genes expressed in at least one cell were retained. Preprocessing, normalization and scaling of the data were carried out using inbuilt Seurat functions. Later, a graph-based clustering approach was used to identify subpopulations of cells and also to optimize the clustering approach to get finalized clusters at resolution 0.5. A list of markers genes for these clusters was obtained and used to further characterize the subpopulations. Furthermore, for the GSE117783 dataset, we analysed the crypts and whole epithelial cells separately. In the crypts sub-dataset, we merged all of the irradiated and normal cells from different clusters together and carried out differential expression analysis using Wilcox tests between the irradiated (cells in bulk) versus normal (cells in bulk) condition. For all the graphs we used the inbuilt functions in Seurat v.3.1.3.
Comparison of the bulk DSS kinetics dataset and scRNA-seq data from irradiated crypts
Time-series bulk RNA-seq data GSE131032 (ref. 5) were downloaded from the GEO. In this dataset, published by our group, we determined in an unbiased manner which genes and pathways are differentially regulated during mouse colonic inflammation followed by tissue regeneration. From this publication, we obtained supplementary dataset 1, containing all the DEGs, which were compared with the upregulated genes in irradiated crypts cells in bulk using jvenn program68. Functional enrichment analysis using enrichR (v.2.1)69 was performed on the genes shared between these two datasets (that is, DEGs in DSS kinetics and upregulated genes in irradiated crypts compared to control). Graphical plotting was performed using inbuilt functions in R.
Analysis of the CRC microarray dataset GSE39582
Analysis of the CRC microarray dataset GSE39582 (ref. 47) was done in R. Probes with a log2 signal below 5 in at least 3 samples were filtered out. Data were quantile normalized and processed for differential gene expression using limma package70 by testing each CRC subgroup to the healthy control group. Genes with FDRâ<â0.01 and fold changeâ>â1.5 were considered to be significant. Samples were stratified in to six tumour subtypes (c1 to c6) based on clinicopathological and molecular differences used in the published study.
Statistical analysis
All statistical analysis, unless otherwise indicated, were performed using GraphPad Prism 9, v.9.5.0. No statistical methods were used to predetermine sample size. Details of statistical tests are provided in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.